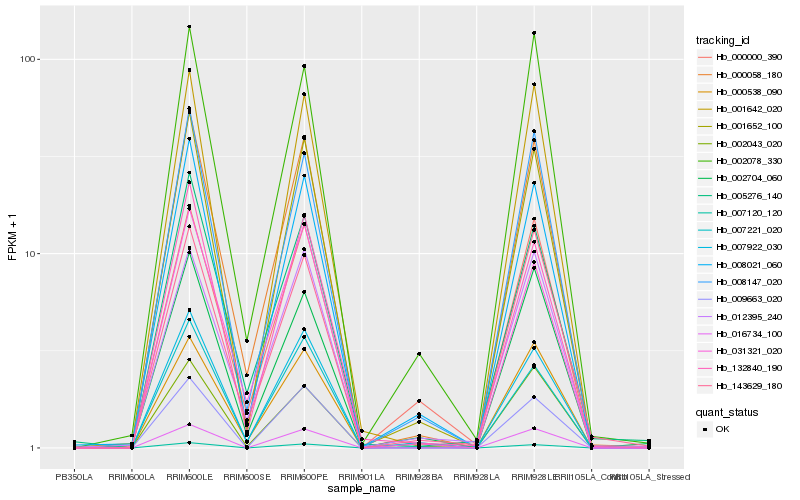
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000058_180 |
0.0 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 2 |
Hb_001652_100 |
0.055518444 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 3 |
Hb_007922_030 |
0.0624962905 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 4 |
Hb_000538_090 |
0.0633769436 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1603380 [Ricinus communis] |
| 5 |
Hb_008147_020 |
0.0644435679 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 6 |
Hb_008021_060 |
0.0645155063 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 7 |
Hb_001642_020 |
0.0772484317 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 8 |
Hb_007221_020 |
0.0782133596 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_031321_020 |
0.0805050445 |
- |
- |
hypothetical protein POPTR_0003s15520g [Populus trichocarpa] |
| 10 |
Hb_016734_100 |
0.08185852 |
- |
- |
- |
| 11 |
Hb_007120_120 |
0.0820260316 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_002043_020 |
0.0825400465 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 13 |
Hb_132840_190 |
0.0830911528 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_005276_140 |
0.083228283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002704_060 |
0.0833923202 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 16 |
Hb_143629_180 |
0.0836746689 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 17 |
Hb_002078_330 |
0.0844968182 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 18 |
Hb_012395_240 |
0.0854275566 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 19 |
Hb_009663_020 |
0.0863782766 |
- |
- |
- |
| 20 |
Hb_000000_390 |
0.0882303086 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |