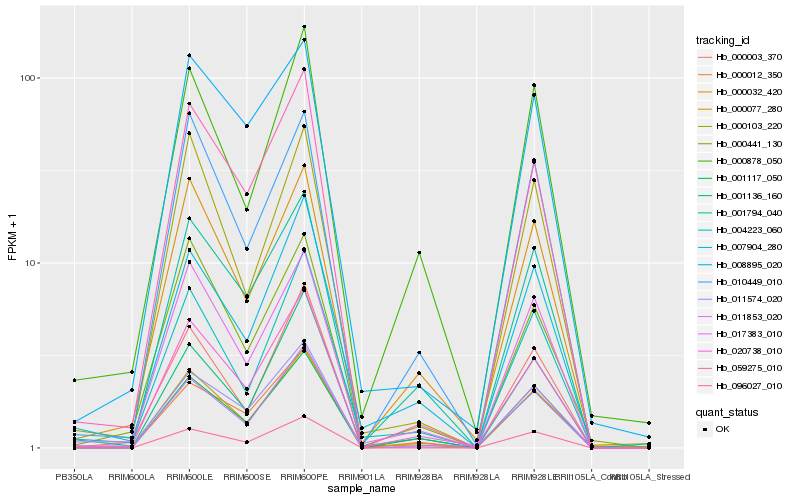
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_280 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_017383_010 |
0.0679719496 |
- |
- |
PREDICTED: uncharacterized protein LOC105628176 [Jatropha curcas] |
| 3 |
Hb_004223_060 |
0.0807626134 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 4 |
Hb_020738_010 |
0.0808700991 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 5 |
Hb_000032_420 |
0.085214525 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 6 |
Hb_007904_280 |
0.0853871854 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 7 |
Hb_000103_220 |
0.0904450024 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |
| 8 |
Hb_001794_040 |
0.0906863619 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 9 |
Hb_011853_020 |
0.0915149782 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 10 |
Hb_000003_370 |
0.0934900159 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 11 |
Hb_000441_130 |
0.0945019218 |
- |
- |
PREDICTED: homeobox-leucine zipper protein HOX32 [Vitis vinifera] |
| 12 |
Hb_000878_050 |
0.0955244656 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 13 |
Hb_059275_010 |
0.0970296997 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 14 |
Hb_010449_010 |
0.0970813353 |
- |
- |
l-ascorbate oxidase, putative [Ricinus communis] |
| 15 |
Hb_001117_050 |
0.0995484414 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 16 |
Hb_000012_350 |
0.0999811467 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 17 |
Hb_096027_010 |
0.1013477058 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_001136_160 |
0.1021891987 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_008895_020 |
0.1040295432 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 20 |
Hb_011574_020 |
0.1048402573 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |