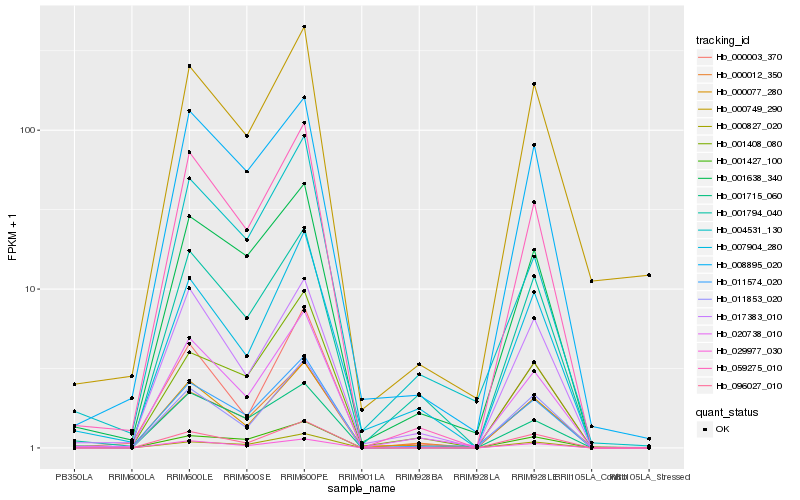
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011574_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 2 |
Hb_020738_010 |
0.0658468073 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 3 |
Hb_001638_340 |
0.0689931213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_059275_010 |
0.0750745411 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 5 |
Hb_000012_350 |
0.0752262981 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 6 |
Hb_008895_020 |
0.1015563956 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 7 |
Hb_001794_040 |
0.1027894548 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000077_280 |
0.1048402573 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_011853_020 |
0.1084014089 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 10 |
Hb_096027_010 |
0.1111984622 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001408_080 |
0.1113292353 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 12 |
Hb_029977_030 |
0.1113742618 |
transcription factor |
TF Family: bZIP |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001427_100 |
0.1127633823 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 14 |
Hb_017383_010 |
0.1138401213 |
- |
- |
PREDICTED: uncharacterized protein LOC105628176 [Jatropha curcas] |
| 15 |
Hb_000003_370 |
0.1141952055 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 16 |
Hb_007904_280 |
0.1155115261 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 17 |
Hb_000749_290 |
0.1171694599 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 18 |
Hb_001715_060 |
0.1174068425 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |
| 19 |
Hb_004531_130 |
0.1182405991 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000827_020 |
0.1184918883 |
- |
- |
unnamed protein product [Coffea canephora] |