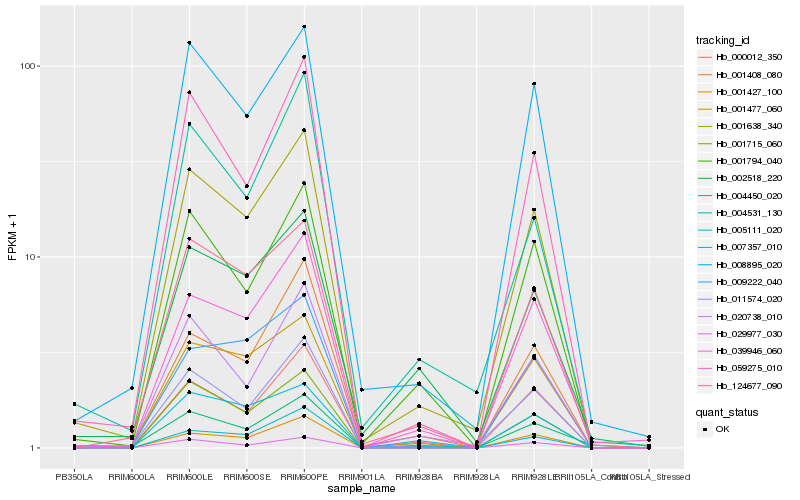
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001638_340 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011574_020 |
0.0689931213 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 3 |
Hb_008895_020 |
0.0794364703 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 4 |
Hb_124677_090 |
0.0857937818 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001477_060 |
0.0867991374 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 6 |
Hb_000012_350 |
0.0904955742 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 7 |
Hb_059275_010 |
0.0905388942 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 8 |
Hb_020738_010 |
0.0918363577 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 9 |
Hb_001715_060 |
0.0929775132 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |
| 10 |
Hb_001427_100 |
0.0963651938 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 11 |
Hb_001794_040 |
0.0964887861 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 12 |
Hb_039946_060 |
0.1040144349 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 13 |
Hb_009222_040 |
0.1088030391 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 14 |
Hb_001408_080 |
0.112209855 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 15 |
Hb_007357_010 |
0.1147297409 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_029977_030 |
0.1164431231 |
transcription factor |
TF Family: bZIP |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004531_130 |
0.1184574508 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_002518_220 |
0.1194351225 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004450_020 |
0.1196453714 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_005111_020 |
0.12015114 |
- |
- |
hypothetical protein CISIN_1g0417821mg, partial [Citrus sinensis] |