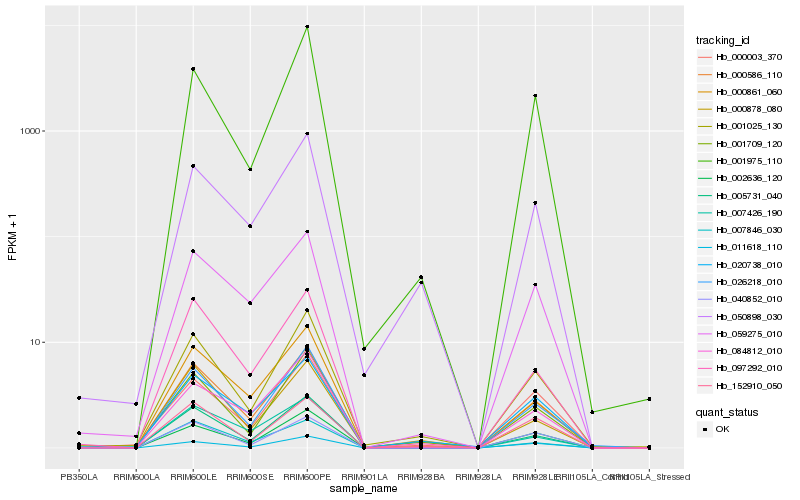
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000586_110 |
0.0 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 2 |
Hb_026218_010 |
0.066307975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005731_040 |
0.06672156 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 4 |
Hb_097292_010 |
0.0776341345 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 5 |
Hb_000861_060 |
0.079811751 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 6 |
Hb_001025_130 |
0.0821959767 |
- |
- |
PREDICTED: uncharacterized protein LOC105640342 [Jatropha curcas] |
| 7 |
Hb_002636_120 |
0.087711017 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 8 |
Hb_059275_010 |
0.0902669572 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 9 |
Hb_001709_120 |
0.092925763 |
- |
- |
hypothetical protein RCOM_1313640 [Ricinus communis] |
| 10 |
Hb_040852_010 |
0.0998253053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_084812_010 |
0.1045438379 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21B [Vitis vinifera] |
| 12 |
Hb_020738_010 |
0.1052764232 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 13 |
Hb_007846_030 |
0.1093432869 |
- |
- |
Uncharacterized protein TCM_021839 [Theobroma cacao] |
| 14 |
Hb_007426_190 |
0.1100589757 |
- |
- |
PREDICTED: uncharacterized protein LOC103325200 [Prunus mume] |
| 15 |
Hb_050898_030 |
0.1112054219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000878_080 |
0.1115464018 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 17 |
Hb_011618_110 |
0.1119810696 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 18 |
Hb_001975_110 |
0.1145009766 |
- |
- |
- |
| 19 |
Hb_000003_370 |
0.1154351606 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 20 |
Hb_152910_050 |
0.1181440449 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |