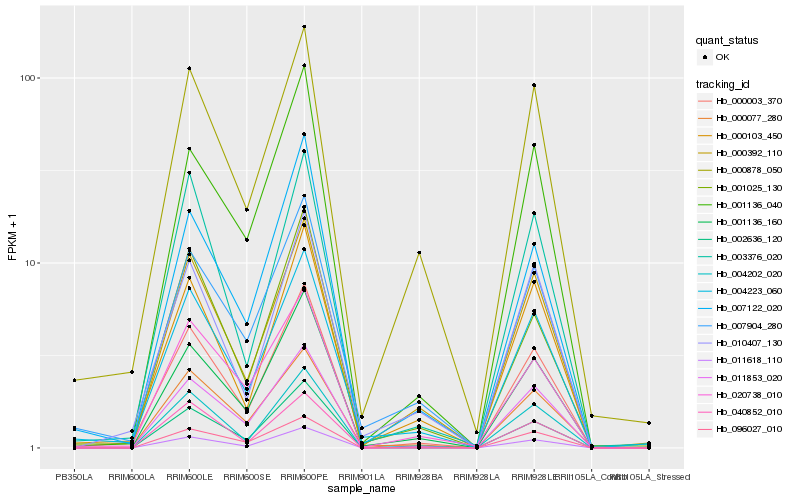
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_370 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 2 |
Hb_011618_110 |
0.0752326554 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 3 |
Hb_011853_020 |
0.0766290906 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 4 |
Hb_004202_020 |
0.0786827546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002636_120 |
0.0798976718 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 6 |
Hb_001136_160 |
0.0813594941 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_004223_060 |
0.081800464 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 8 |
Hb_000103_450 |
0.0866616045 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 9 |
Hb_007122_020 |
0.0912759155 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 10 |
Hb_001136_040 |
0.0923334993 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_000077_280 |
0.0934900159 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_001025_130 |
0.0965256941 |
- |
- |
PREDICTED: uncharacterized protein LOC105640342 [Jatropha curcas] |
| 13 |
Hb_003376_020 |
0.0967327375 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_096027_010 |
0.0977165911 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000392_110 |
0.0977786126 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_010407_130 |
0.0981386971 |
- |
- |
hypothetical protein POPTR_0008s16680g [Populus trichocarpa] |
| 17 |
Hb_020738_010 |
0.0982887968 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 18 |
Hb_000878_050 |
0.0988924417 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 19 |
Hb_040852_010 |
0.0990759195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007904_280 |
0.0999325981 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |