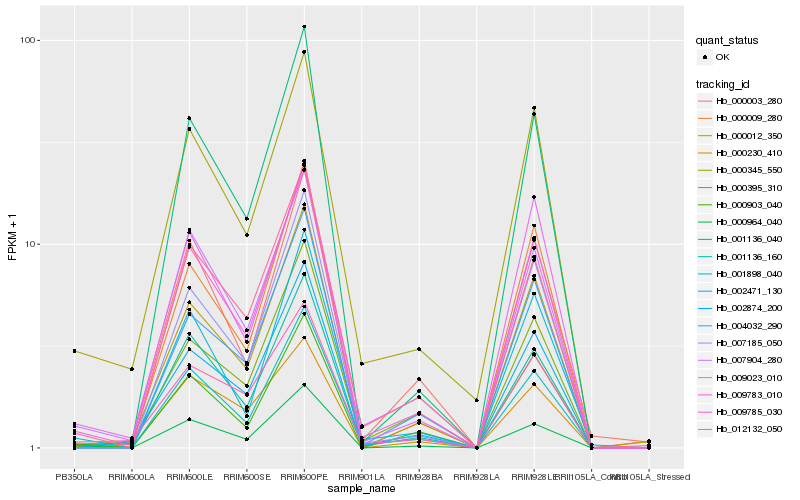
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009783_010 |
0.0 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 2 |
Hb_007904_280 |
0.0639041243 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 3 |
Hb_001898_040 |
0.0663515427 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 4 |
Hb_012132_050 |
0.066873017 |
- |
- |
PREDICTED: aspartic proteinase PCS1 [Jatropha curcas] |
| 5 |
Hb_001136_040 |
0.0671876106 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_001136_160 |
0.0723240347 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_007185_050 |
0.07286825 |
- |
- |
hypothetical protein JCGZ_22475 [Jatropha curcas] |
| 8 |
Hb_000230_410 |
0.0763403143 |
- |
- |
PREDICTED: uncharacterized protein LOC105631891 [Jatropha curcas] |
| 9 |
Hb_002874_200 |
0.0780164655 |
- |
- |
PREDICTED: uncharacterized protein LOC105109832 [Populus euphratica] |
| 10 |
Hb_000964_040 |
0.080116879 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER isoform X2 [Jatropha curcas] |
| 11 |
Hb_004032_290 |
0.085748521 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_000395_310 |
0.0873470417 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 13 |
Hb_002471_130 |
0.0877521534 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |
| 14 |
Hb_000009_280 |
0.0898748495 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 15 |
Hb_000345_550 |
0.0901915824 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 16 |
Hb_000903_040 |
0.0905447835 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 17 |
Hb_009785_030 |
0.0909341147 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 18 |
Hb_009023_010 |
0.0917773705 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 19 |
Hb_000012_350 |
0.0934998108 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 20 |
Hb_000003_280 |
0.0948841847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |