| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
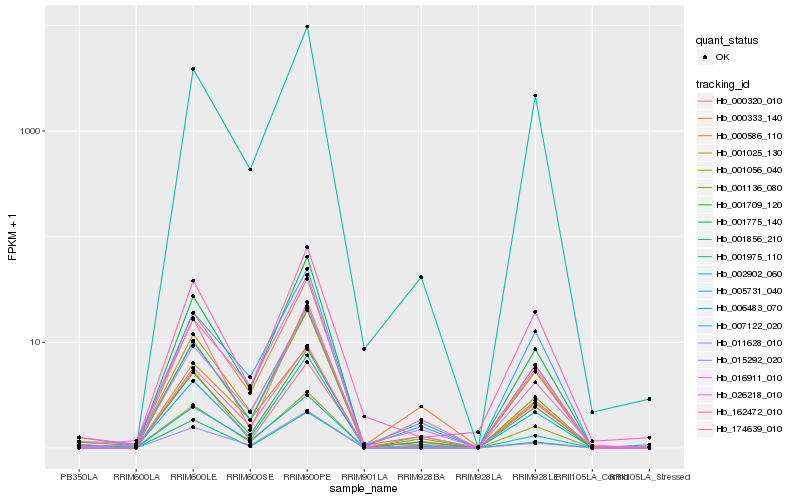
Hb_001709_120 |
0.0 |
- |
- |
hypothetical protein RCOM_1313640 [Ricinus communis] |
| 2 |
Hb_001025_130 |
0.0513808233 |
- |
- |
PREDICTED: uncharacterized protein LOC105640342 [Jatropha curcas] |
| 3 |
Hb_015292_020 |
0.0760361235 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_002902_060 |
0.0761271903 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 5 |
Hb_174639_010 |
0.0801661821 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_001056_040 |
0.0845146335 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 7 |
Hb_006483_070 |
0.0845850747 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 8 |
Hb_001975_110 |
0.0853415545 |
- |
- |
- |
| 9 |
Hb_001136_080 |
0.0858150207 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 10 |
Hb_000333_140 |
0.0920704453 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 11 |
Hb_000586_110 |
0.092925763 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 12 |
Hb_026218_010 |
0.0963277648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_162472_010 |
0.0993088659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011628_010 |
0.1014720833 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 15 |
Hb_001856_210 |
0.1029316712 |
- |
- |
PREDICTED: uncharacterized protein LOC105644171 [Jatropha curcas] |
| 16 |
Hb_001775_140 |
0.1044343557 |
- |
- |
Phytosulfokine 3 precursor, putative isoform 2 [Theobroma cacao] |
| 17 |
Hb_000320_010 |
0.1054133438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005731_040 |
0.1055046618 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 19 |
Hb_007122_020 |
0.1059987331 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 20 |
Hb_016911_010 |
0.1086240059 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |