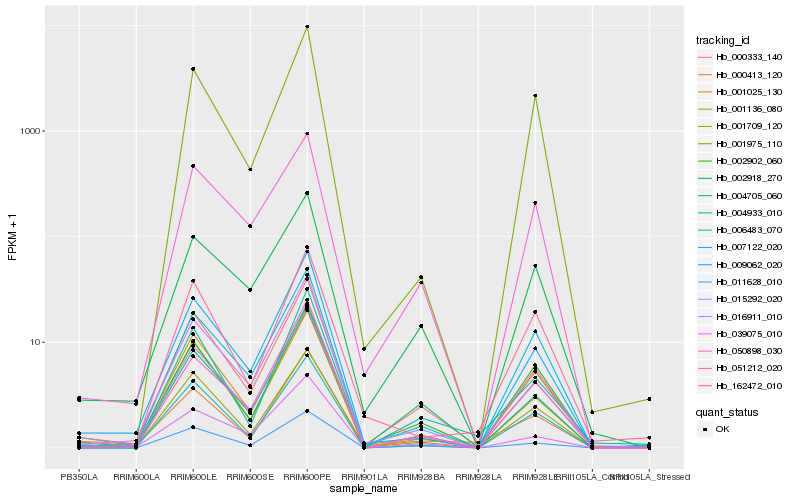
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015292_020 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000333_140 |
0.0690450414 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 3 |
Hb_001709_120 |
0.0760361235 |
- |
- |
hypothetical protein RCOM_1313640 [Ricinus communis] |
| 4 |
Hb_001025_130 |
0.0761085528 |
- |
- |
PREDICTED: uncharacterized protein LOC105640342 [Jatropha curcas] |
| 5 |
Hb_002902_060 |
0.0799118676 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 6 |
Hb_016911_010 |
0.0822428949 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 7 |
Hb_007122_020 |
0.087004687 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 8 |
Hb_001136_080 |
0.0897529401 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 9 |
Hb_009062_020 |
0.0900396905 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |
| 10 |
Hb_050898_030 |
0.0917736693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002918_270 |
0.0927507777 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 12 |
Hb_001975_110 |
0.094136894 |
- |
- |
- |
| 13 |
Hb_011628_010 |
0.0946710985 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 14 |
Hb_051212_020 |
0.0950414177 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 15 |
Hb_006483_070 |
0.0992922272 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 16 |
Hb_000413_120 |
0.1008297741 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 17 |
Hb_162472_010 |
0.1067262399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004933_010 |
0.1068873442 |
- |
- |
hypothetical protein POPTR_0018s10810g [Populus trichocarpa] |
| 19 |
Hb_039075_010 |
0.1090970403 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 20 |
Hb_004705_060 |
0.1108911919 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |