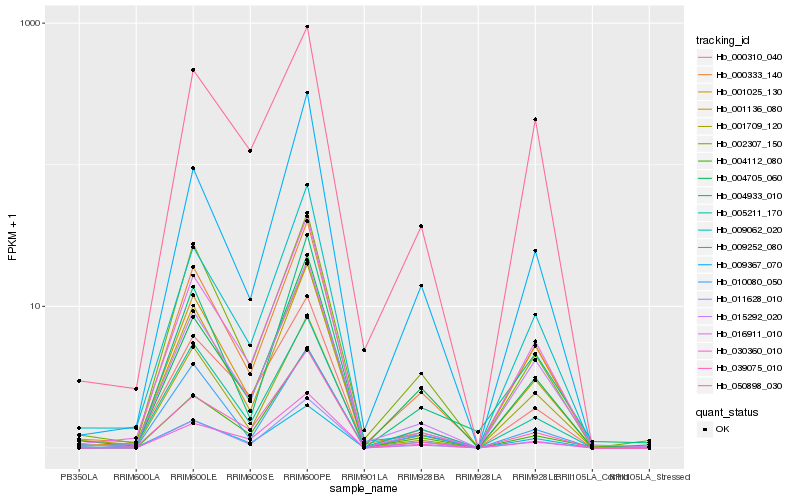
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000333_140 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 2 |
Hb_011628_010 |
0.0575375778 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 3 |
Hb_005211_170 |
0.0680597161 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 4 |
Hb_001136_080 |
0.0681144067 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 5 |
Hb_015292_020 |
0.0690450414 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_002307_150 |
0.0708305335 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 7 |
Hb_000310_040 |
0.0824597173 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 8 |
Hb_004705_060 |
0.0860240823 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 9 |
Hb_016911_010 |
0.0897788585 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 10 |
Hb_039075_010 |
0.0914528503 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 11 |
Hb_001709_120 |
0.0920704453 |
- |
- |
hypothetical protein RCOM_1313640 [Ricinus communis] |
| 12 |
Hb_009367_070 |
0.0996908787 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 13 |
Hb_009062_020 |
0.1012159656 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |
| 14 |
Hb_030360_010 |
0.1032081358 |
- |
- |
- |
| 15 |
Hb_009252_080 |
0.10347214 |
- |
- |
Polygalacturonase-1 non-catalytic subunit beta precursor, putative [Ricinus communis] |
| 16 |
Hb_004933_010 |
0.1056643983 |
- |
- |
hypothetical protein POPTR_0018s10810g [Populus trichocarpa] |
| 17 |
Hb_001025_130 |
0.1061028501 |
- |
- |
PREDICTED: uncharacterized protein LOC105640342 [Jatropha curcas] |
| 18 |
Hb_004112_080 |
0.1061309569 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 19 |
Hb_010080_050 |
0.1082798738 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 20 |
Hb_050898_030 |
0.1084190225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |