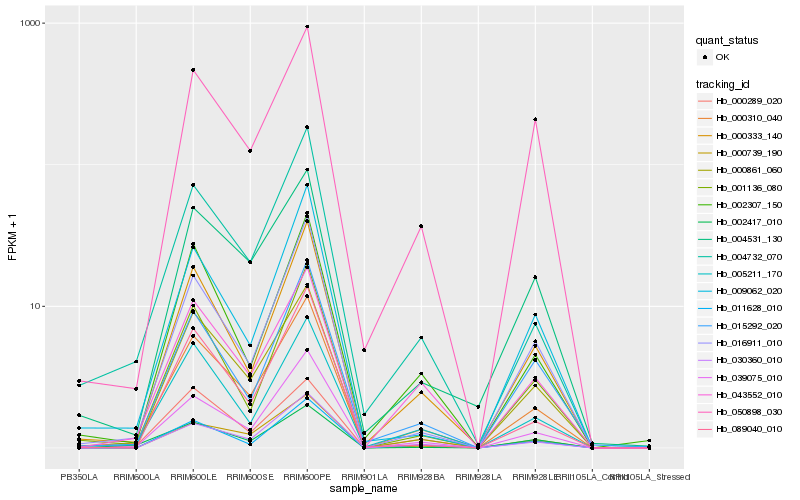
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000310_040 |
0.0 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 2 |
Hb_039075_010 |
0.0750909674 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 3 |
Hb_000333_140 |
0.0824597173 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 4 |
Hb_005211_170 |
0.0847986187 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 5 |
Hb_030360_010 |
0.0873562964 |
- |
- |
- |
| 6 |
Hb_004732_070 |
0.0921942921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000861_060 |
0.0954207118 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 8 |
Hb_043552_010 |
0.0954227112 |
- |
- |
PREDICTED: pectinesterase/pectinesterase inhibitor PPE8B isoform X1 [Jatropha curcas] |
| 9 |
Hb_000739_190 |
0.0975962466 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_016911_010 |
0.1012897379 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 11 |
Hb_002307_150 |
0.1015556018 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 12 |
Hb_011628_010 |
0.1029357195 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 13 |
Hb_001136_080 |
0.107826502 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 14 |
Hb_089040_010 |
0.1131148314 |
- |
- |
Glycosyl hydrolase family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_002417_010 |
0.1137784577 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 16 |
Hb_004531_130 |
0.1149098645 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_050898_030 |
0.1155833607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_015292_020 |
0.1184336646 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000289_020 |
0.1194320937 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 20 |
Hb_009062_020 |
0.1194640206 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |