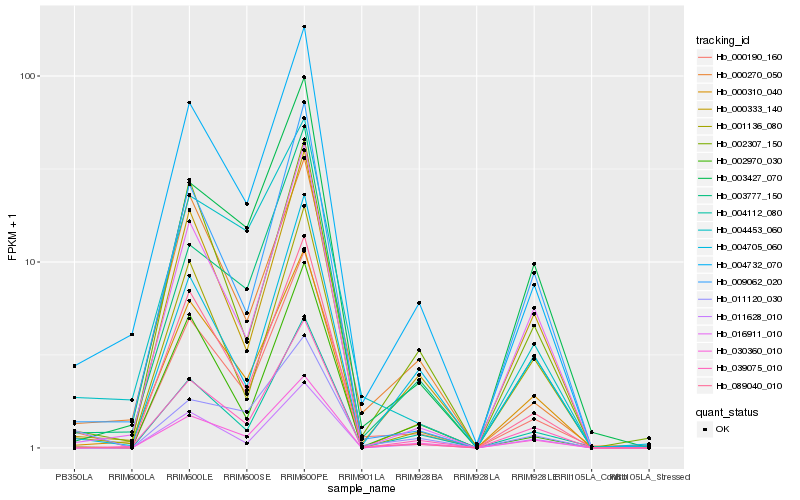
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000190_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649940 isoform X2 [Jatropha curcas] |
| 2 |
Hb_004732_070 |
0.0969182542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_089040_010 |
0.1166392025 |
- |
- |
Glycosyl hydrolase family protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_004112_080 |
0.1214354212 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 5 |
Hb_039075_010 |
0.1291040174 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 6 |
Hb_000310_040 |
0.1298669655 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 7 |
Hb_030360_010 |
0.1299272392 |
- |
- |
- |
| 8 |
Hb_003777_150 |
0.1330803923 |
- |
- |
PREDICTED: uncharacterized protein LOC105640938 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002970_030 |
0.1338636553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000333_140 |
0.1355187774 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 11 |
Hb_000270_050 |
0.136077358 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 12 |
Hb_009062_020 |
0.1391712476 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |
| 13 |
Hb_002307_150 |
0.1397024079 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 14 |
Hb_004453_060 |
0.1425588551 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2 [Jatropha curcas] |
| 15 |
Hb_003427_070 |
0.1446178677 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 16 |
Hb_001136_080 |
0.1446781259 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 17 |
Hb_016911_010 |
0.1452434124 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 18 |
Hb_011628_010 |
0.1490632094 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 19 |
Hb_004705_060 |
0.1510518973 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 20 |
Hb_011120_030 |
0.1513048018 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |