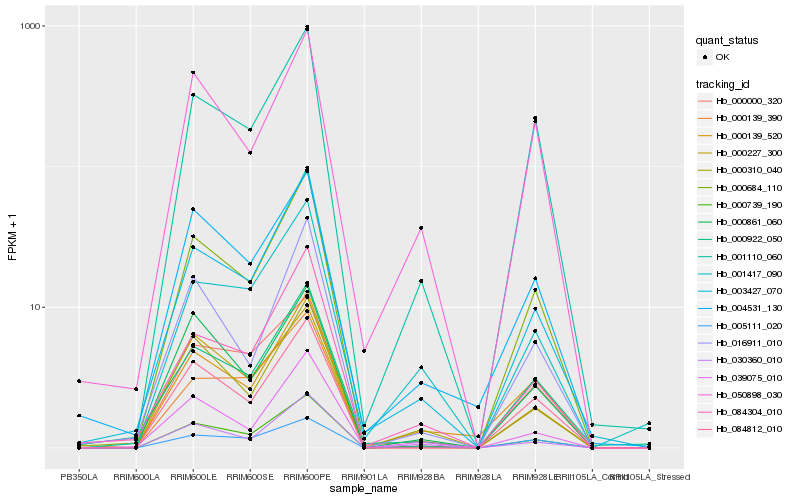
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000739_190 |
0.0 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_003427_070 |
0.078597033 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 3 |
Hb_030360_010 |
0.0805040942 |
- |
- |
- |
| 4 |
Hb_000684_110 |
0.0858759881 |
- |
- |
hypothetical protein POPTR_0006s13250g [Populus trichocarpa] |
| 5 |
Hb_039075_010 |
0.0925857468 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 6 |
Hb_000310_040 |
0.0975962466 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 7 |
Hb_001110_060 |
0.101403619 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 8 |
Hb_000139_520 |
0.1040404182 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 9 |
Hb_084304_010 |
0.1046088758 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X2 [Jatropha curcas] |
| 10 |
Hb_084812_010 |
0.1056034338 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21B [Vitis vinifera] |
| 11 |
Hb_005111_020 |
0.1059406442 |
- |
- |
hypothetical protein CISIN_1g0417821mg, partial [Citrus sinensis] |
| 12 |
Hb_001417_090 |
0.1059574574 |
- |
- |
PREDICTED: uncharacterized protein LOC105650625 [Jatropha curcas] |
| 13 |
Hb_000000_320 |
0.1077721123 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000861_060 |
0.1082070245 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 15 |
Hb_004531_130 |
0.1128987201 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000227_300 |
0.1163902551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000922_050 |
0.1179671385 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 18 |
Hb_000139_390 |
0.1189524271 |
- |
- |
- |
| 19 |
Hb_016911_010 |
0.1192524574 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 20 |
Hb_050898_030 |
0.1215840941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |