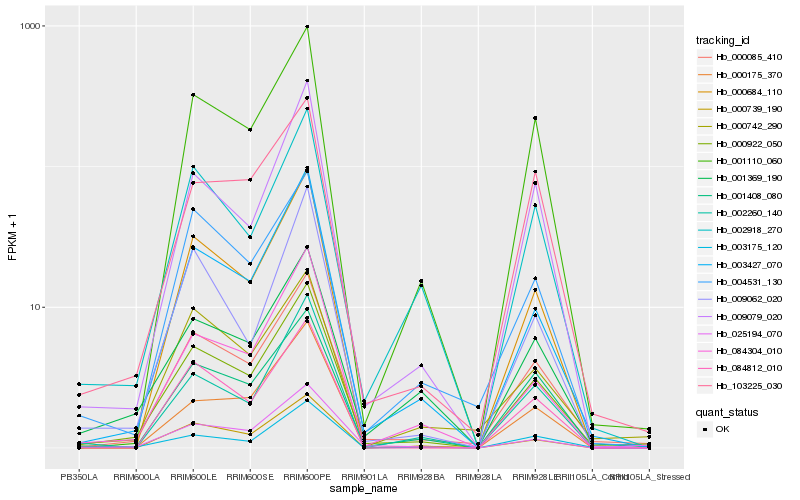
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_050 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 2 |
Hb_000085_410 |
0.0606122782 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 3 |
Hb_003427_070 |
0.0852875462 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 4 |
Hb_001110_060 |
0.1030614857 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 5 |
Hb_000684_110 |
0.1086565049 |
- |
- |
hypothetical protein POPTR_0006s13250g [Populus trichocarpa] |
| 6 |
Hb_003175_120 |
0.1093160923 |
- |
- |
PREDICTED: uncharacterized protein LOC105141472 isoform X1 [Populus euphratica] |
| 7 |
Hb_025194_070 |
0.1114735525 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4-like [Jatropha curcas] |
| 8 |
Hb_084304_010 |
0.1115991262 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X2 [Jatropha curcas] |
| 9 |
Hb_009079_020 |
0.1118066671 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 10 |
Hb_001408_080 |
0.1119624345 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 11 |
Hb_001369_190 |
0.1140065946 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 12 |
Hb_002918_270 |
0.1176421565 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 13 |
Hb_000739_190 |
0.1179671385 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_084812_010 |
0.1221195706 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21B [Vitis vinifera] |
| 15 |
Hb_103225_030 |
0.1252884371 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000742_290 |
0.1253303444 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 17 |
Hb_000175_370 |
0.1257192941 |
- |
- |
hypothetical protein EUGRSUZ_F03862 [Eucalyptus grandis] |
| 18 |
Hb_002260_140 |
0.1271721696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004531_130 |
0.1284650559 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_009062_020 |
0.1285691252 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |