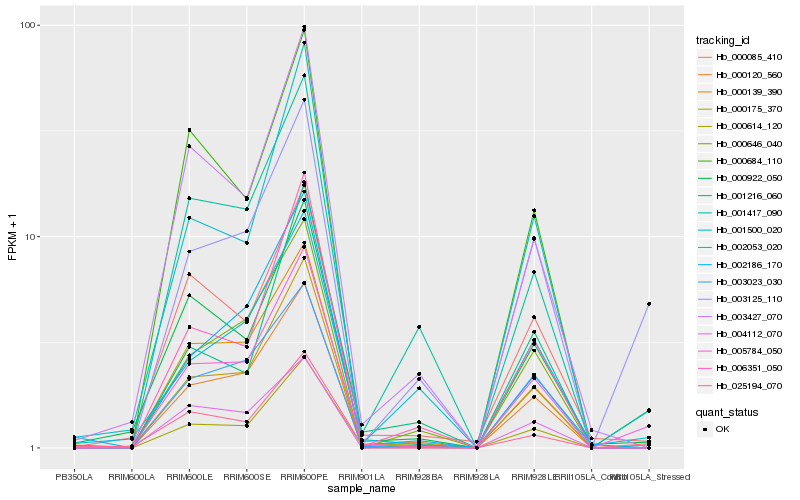
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005784_050 |
0.0 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_002053_020 |
0.1133366949 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 3 |
Hb_000175_370 |
0.1161625727 |
- |
- |
hypothetical protein EUGRSUZ_F03862 [Eucalyptus grandis] |
| 4 |
Hb_025194_070 |
0.1209916887 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4-like [Jatropha curcas] |
| 5 |
Hb_003125_110 |
0.121239298 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 6 |
Hb_000120_560 |
0.1236674669 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_000646_040 |
0.1320387269 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 8 |
Hb_000139_390 |
0.1333079195 |
- |
- |
- |
| 9 |
Hb_000922_050 |
0.1355997926 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 10 |
Hb_000614_120 |
0.1441636715 |
- |
- |
hypothetical protein JCGZ_17923 [Jatropha curcas] |
| 11 |
Hb_001417_090 |
0.1448488154 |
- |
- |
PREDICTED: uncharacterized protein LOC105650625 [Jatropha curcas] |
| 12 |
Hb_000085_410 |
0.1450057691 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 13 |
Hb_001216_060 |
0.1472326985 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002186_170 |
0.147925649 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 15 |
Hb_003023_030 |
0.1479871356 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 16 |
Hb_006351_050 |
0.1493539695 |
- |
- |
PREDICTED: uncharacterized protein LOC105133646 [Populus euphratica] |
| 17 |
Hb_003427_070 |
0.150282576 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 18 |
Hb_004112_070 |
0.1507343716 |
- |
- |
- |
| 19 |
Hb_001500_020 |
0.151528271 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000684_110 |
0.1517672221 |
- |
- |
hypothetical protein POPTR_0006s13250g [Populus trichocarpa] |