| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
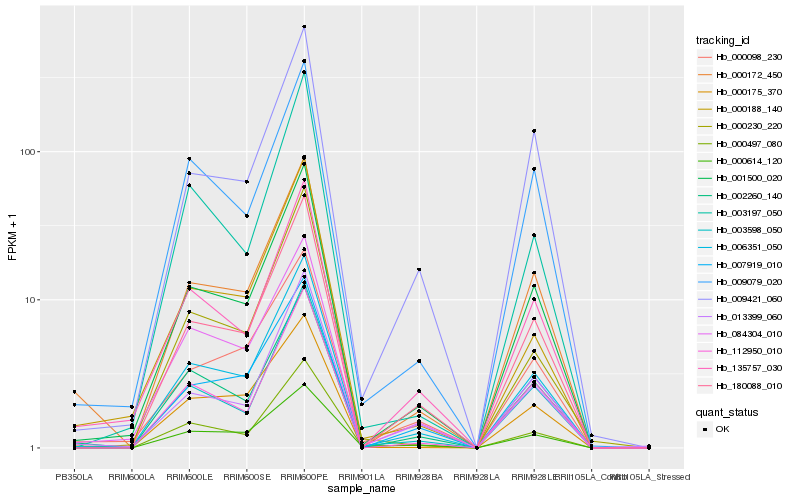
Hb_006351_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105133646 [Populus euphratica] |
| 2 |
Hb_001500_020 |
0.0311812184 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_180088_010 |
0.0420521352 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_003598_050 |
0.0661226821 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000497_080 |
0.0690581859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_112950_010 |
0.0752704312 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 7 |
Hb_007919_010 |
0.0764026652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002260_140 |
0.0791957576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_135757_030 |
0.0803658667 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 10 |
Hb_000172_450 |
0.080586643 |
- |
- |
Uncharacterized protein TCM_001203 [Theobroma cacao] |
| 11 |
Hb_084304_010 |
0.0880078461 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X2 [Jatropha curcas] |
| 12 |
Hb_009079_020 |
0.08866483 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 13 |
Hb_009421_060 |
0.0902214727 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 14 |
Hb_013399_060 |
0.0931769086 |
- |
- |
PREDICTED: uncharacterized protein LOC105133408 [Populus euphratica] |
| 15 |
Hb_000614_120 |
0.094380595 |
- |
- |
hypothetical protein JCGZ_17923 [Jatropha curcas] |
| 16 |
Hb_000175_370 |
0.0964630535 |
- |
- |
hypothetical protein EUGRSUZ_F03862 [Eucalyptus grandis] |
| 17 |
Hb_003197_050 |
0.0965803605 |
- |
- |
hypothetical protein JCGZ_04279 [Jatropha curcas] |
| 18 |
Hb_000098_230 |
0.0967728004 |
- |
- |
Potassium channel AKT2/3, putative [Ricinus communis] |
| 19 |
Hb_000230_220 |
0.0977552564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000188_140 |
0.0990101007 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |