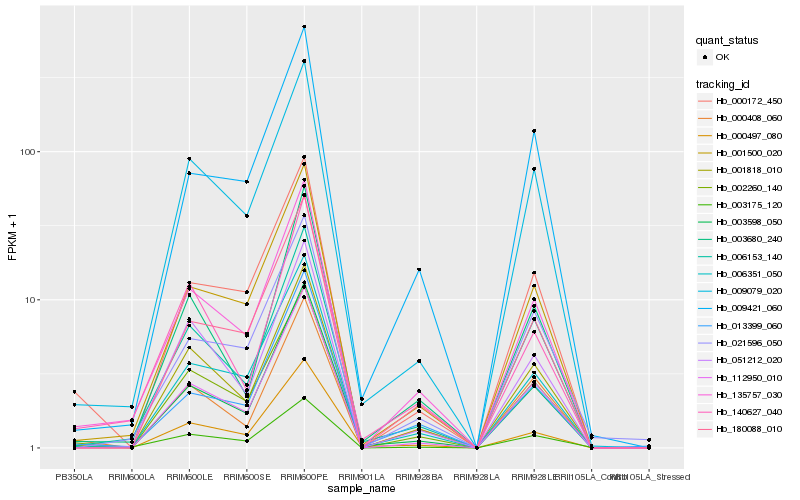
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135757_030 |
0.0 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 2 |
Hb_002260_140 |
0.0675265161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001500_020 |
0.0682804961 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_001818_010 |
0.07595442 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 5 |
Hb_003175_120 |
0.0779975934 |
- |
- |
PREDICTED: uncharacterized protein LOC105141472 isoform X1 [Populus euphratica] |
| 6 |
Hb_003598_050 |
0.0792148888 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 7 |
Hb_006351_050 |
0.0803658667 |
- |
- |
PREDICTED: uncharacterized protein LOC105133646 [Populus euphratica] |
| 8 |
Hb_009079_020 |
0.0820856568 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 9 |
Hb_006153_140 |
0.0829233363 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 10 |
Hb_112950_010 |
0.0883247098 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 11 |
Hb_180088_010 |
0.0895908819 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000172_450 |
0.0949671055 |
- |
- |
Uncharacterized protein TCM_001203 [Theobroma cacao] |
| 13 |
Hb_013399_060 |
0.0981330343 |
- |
- |
PREDICTED: uncharacterized protein LOC105133408 [Populus euphratica] |
| 14 |
Hb_000408_060 |
0.0995040129 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 15 |
Hb_003680_240 |
0.1012138522 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 16 |
Hb_051212_020 |
0.1018322929 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 17 |
Hb_140627_040 |
0.1050153502 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 18 |
Hb_000497_080 |
0.1056447454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_009421_060 |
0.1060978174 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 20 |
Hb_021596_050 |
0.1066949352 |
- |
- |
conserved hypothetical protein [Ricinus communis] |