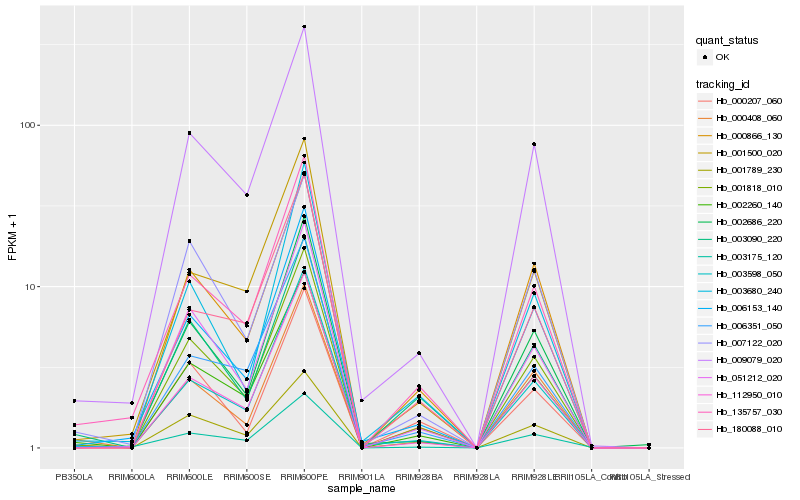
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001818_010 |
0.0 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 2 |
Hb_002260_140 |
0.0478517067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_051212_020 |
0.0553073621 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 4 |
Hb_009079_020 |
0.0672479244 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 5 |
Hb_000866_130 |
0.0746255627 |
- |
- |
hypothetical protein JCGZ_14397 [Jatropha curcas] |
| 6 |
Hb_112950_010 |
0.0755051994 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 7 |
Hb_135757_030 |
0.07595442 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 8 |
Hb_003090_220 |
0.0767330642 |
- |
- |
PREDICTED: copper transporter 4 [Jatropha curcas] |
| 9 |
Hb_006153_140 |
0.077296408 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 10 |
Hb_001789_230 |
0.084921259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000408_060 |
0.0858695059 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 12 |
Hb_002686_220 |
0.0859338883 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 13 |
Hb_003598_050 |
0.0889464733 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000207_060 |
0.0903688719 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 15 |
Hb_003680_240 |
0.0911315548 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 16 |
Hb_007122_020 |
0.0942164405 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 17 |
Hb_001500_020 |
0.0989227943 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_006351_050 |
0.1005090684 |
- |
- |
PREDICTED: uncharacterized protein LOC105133646 [Populus euphratica] |
| 19 |
Hb_003175_120 |
0.1040342619 |
- |
- |
PREDICTED: uncharacterized protein LOC105141472 isoform X1 [Populus euphratica] |
| 20 |
Hb_180088_010 |
0.1046415945 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |