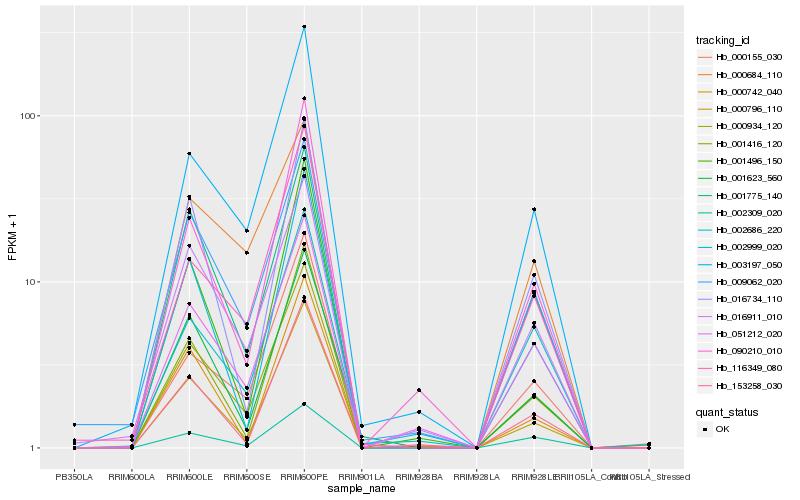
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000934_120 |
0.0 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 2 |
Hb_000796_110 |
0.0736079783 |
- |
- |
PREDICTED: probable pectinesterase 53 [Populus euphratica] |
| 3 |
Hb_001775_140 |
0.0780250802 |
- |
- |
Phytosulfokine 3 precursor, putative isoform 2 [Theobroma cacao] |
| 4 |
Hb_016911_010 |
0.0843528415 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 5 |
Hb_051212_020 |
0.0854898675 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 6 |
Hb_009062_020 |
0.0857443427 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |
| 7 |
Hb_003197_050 |
0.0858843217 |
- |
- |
hypothetical protein JCGZ_04279 [Jatropha curcas] |
| 8 |
Hb_001416_120 |
0.0981055124 |
- |
- |
caspase, putative [Ricinus communis] |
| 9 |
Hb_002309_020 |
0.1011392415 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 10 |
Hb_116349_080 |
0.1013281078 |
- |
- |
Nectarin-1 precursor, putative [Ricinus communis] |
| 11 |
Hb_001623_560 |
0.102397111 |
- |
- |
late embryogenesis abundant protein group 9 protein [Arachis hypogaea] |
| 12 |
Hb_153258_030 |
0.1037688699 |
- |
- |
hypothetical protein RCOM_0853150 [Ricinus communis] |
| 13 |
Hb_000155_030 |
0.1038188994 |
- |
- |
PREDICTED: uncharacterized protein LOC105649591 [Jatropha curcas] |
| 14 |
Hb_016734_110 |
0.104599703 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 15 |
Hb_090210_010 |
0.1075924017 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 16 |
Hb_001496_150 |
0.1106040639 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 17 |
Hb_002999_020 |
0.11094464 |
- |
- |
PREDICTED: ribonuclease 3-like [Jatropha curcas] |
| 18 |
Hb_000684_110 |
0.1139118851 |
- |
- |
hypothetical protein POPTR_0006s13250g [Populus trichocarpa] |
| 19 |
Hb_000742_040 |
0.1156694222 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 20 |
Hb_002686_220 |
0.1157799715 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |