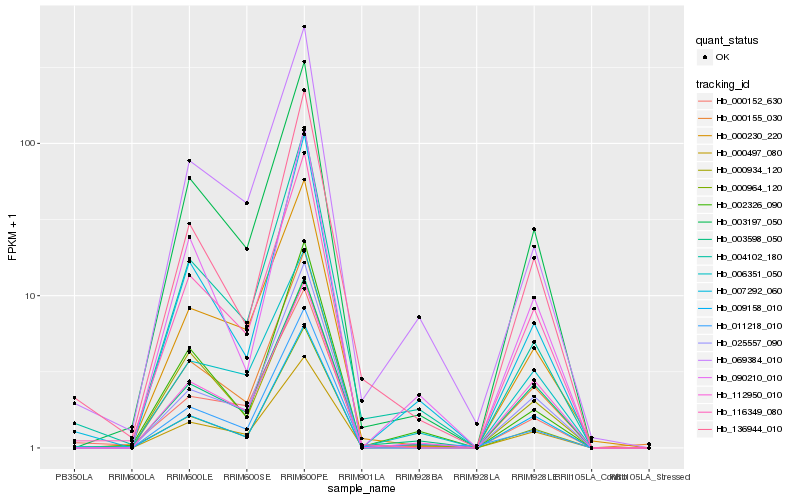
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003197_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_04279 [Jatropha curcas] |
| 2 |
Hb_116349_080 |
0.0424540244 |
- |
- |
Nectarin-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000155_030 |
0.0506783473 |
- |
- |
PREDICTED: uncharacterized protein LOC105649591 [Jatropha curcas] |
| 4 |
Hb_000230_220 |
0.0709577417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000964_120 |
0.079211356 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 6 |
Hb_000497_080 |
0.0795214287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000934_120 |
0.0858843217 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 8 |
Hb_003598_050 |
0.0863851581 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 9 |
Hb_025557_090 |
0.0867947012 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |
| 10 |
Hb_009158_010 |
0.0906539132 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 11 |
Hb_007292_060 |
0.092696252 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |
| 12 |
Hb_136944_010 |
0.0938609059 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 13 |
Hb_090210_010 |
0.0938696011 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 14 |
Hb_004102_180 |
0.094081342 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 15 |
Hb_011218_010 |
0.0950053304 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 16 |
Hb_069384_010 |
0.0955393559 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_112950_010 |
0.0959745417 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 18 |
Hb_006351_050 |
0.0965803605 |
- |
- |
PREDICTED: uncharacterized protein LOC105133646 [Populus euphratica] |
| 19 |
Hb_000152_630 |
0.097132542 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Jatropha curcas] |
| 20 |
Hb_002326_090 |
0.0977255912 |
- |
- |
amino acid transporter, putative [Ricinus communis] |