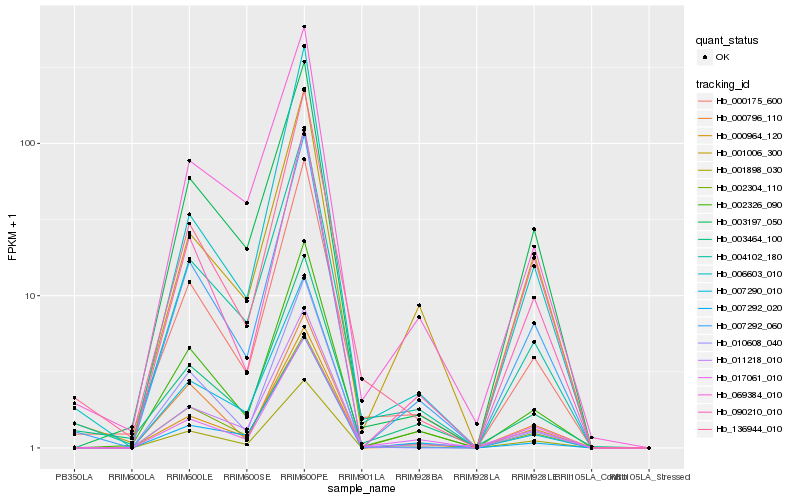
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002326_090 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_007292_060 |
0.0586306958 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |
| 3 |
Hb_000175_600 |
0.0600365091 |
- |
- |
PREDICTED: TATA-binding protein-associated factor 2N isoform X2 [Jatropha curcas] |
| 4 |
Hb_000964_120 |
0.0701513213 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 5 |
Hb_090210_010 |
0.0717043747 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 6 |
Hb_004102_180 |
0.0739565276 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 7 |
Hb_001898_030 |
0.0809271371 |
- |
- |
protein kinase-coding resistance protein [Nicotiana repanda] |
| 8 |
Hb_010608_040 |
0.0851871976 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 9 |
Hb_011218_010 |
0.0858476462 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 10 |
Hb_069384_010 |
0.0876160519 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_003197_050 |
0.0977255912 |
- |
- |
hypothetical protein JCGZ_04279 [Jatropha curcas] |
| 12 |
Hb_007290_010 |
0.0990182805 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_007292_020 |
0.1002678319 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 14 |
Hb_000796_110 |
0.1017336311 |
- |
- |
PREDICTED: probable pectinesterase 53 [Populus euphratica] |
| 15 |
Hb_017061_010 |
0.1051098889 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 16 |
Hb_001006_300 |
0.1083670565 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 17 |
Hb_136944_010 |
0.1087321926 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 18 |
Hb_003464_100 |
0.1094397301 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 19 |
Hb_002304_110 |
0.1104021407 |
- |
- |
hypothetical protein MIMGU_mgv1a023246mg [Erythranthe guttata] |
| 20 |
Hb_006603_010 |
0.111057775 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |