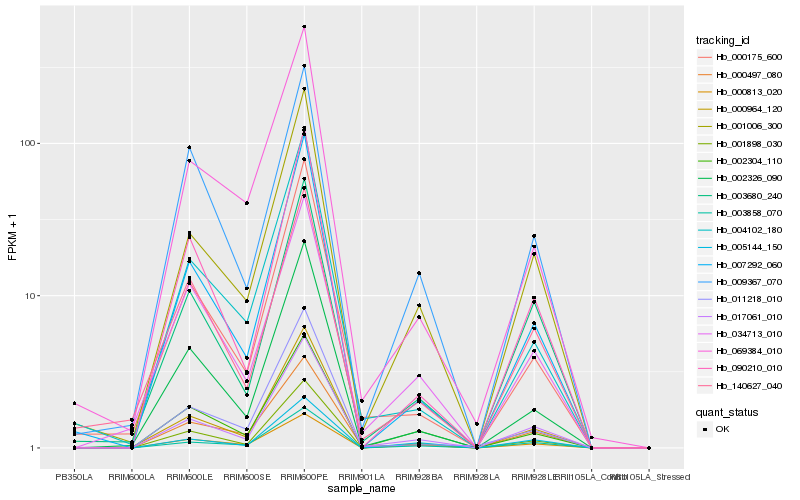
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001898_030 |
0.0 |
- |
- |
protein kinase-coding resistance protein [Nicotiana repanda] |
| 2 |
Hb_002304_110 |
0.0598066038 |
- |
- |
hypothetical protein MIMGU_mgv1a023246mg [Erythranthe guttata] |
| 3 |
Hb_017061_010 |
0.0679855111 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 4 |
Hb_001006_300 |
0.0712313979 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 5 |
Hb_090210_010 |
0.0802528748 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 6 |
Hb_002326_090 |
0.0809271371 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_007292_060 |
0.0840810667 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |
| 8 |
Hb_003858_070 |
0.0848872454 |
- |
- |
PREDICTED: uncharacterized protein LOC105647879 [Jatropha curcas] |
| 9 |
Hb_000964_120 |
0.08674134 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 10 |
Hb_005144_150 |
0.0875346331 |
- |
- |
hypothetical protein VITISV_013572 [Vitis vinifera] |
| 11 |
Hb_000813_020 |
0.0937811477 |
- |
- |
PREDICTED: metacaspase-3-like [Jatropha curcas] |
| 12 |
Hb_009367_070 |
0.0938469988 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 13 |
Hb_034713_010 |
0.095245544 |
- |
- |
PREDICTED: uncharacterized protein LOC105133378 [Populus euphratica] |
| 14 |
Hb_003680_240 |
0.1092846477 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 15 |
Hb_000175_600 |
0.1137987821 |
- |
- |
PREDICTED: TATA-binding protein-associated factor 2N isoform X2 [Jatropha curcas] |
| 16 |
Hb_000497_080 |
0.1162696977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_011218_010 |
0.1180116924 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 18 |
Hb_140627_040 |
0.1193606748 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 19 |
Hb_069384_010 |
0.1197072078 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_004102_180 |
0.121677709 |
- |
- |
zinc finger family protein [Populus trichocarpa] |