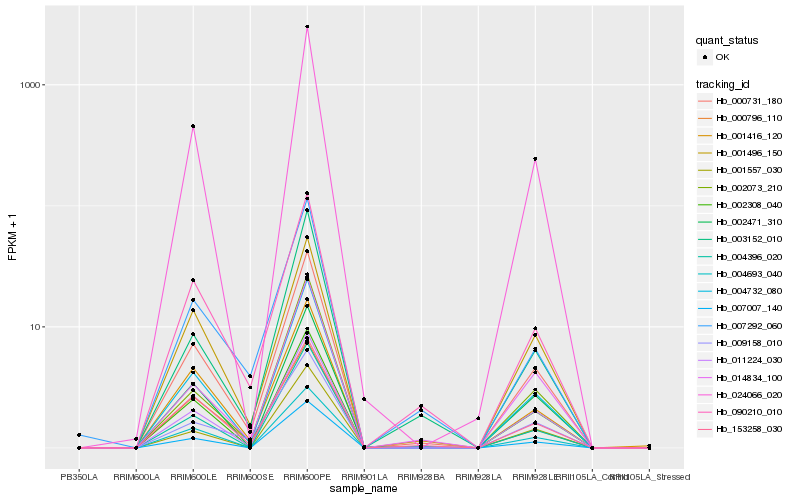
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000731_180 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s01820g [Populus trichocarpa] |
| 2 |
Hb_004732_080 |
0.0317949351 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 3 |
Hb_090210_010 |
0.0597488238 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 4 |
Hb_011224_030 |
0.0660982976 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_007007_140 |
0.0674176433 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL6 [Jatropha curcas] |
| 6 |
Hb_024066_020 |
0.0683031838 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 7 |
Hb_002308_040 |
0.0702038227 |
- |
- |
methyladenine glycosylase family protein [Populus trichocarpa] |
| 8 |
Hb_153258_030 |
0.0725258445 |
- |
- |
hypothetical protein RCOM_0853150 [Ricinus communis] |
| 9 |
Hb_004396_020 |
0.0744786185 |
- |
- |
SAUR-like auxin-responsive protein family [Theobroma cacao] |
| 10 |
Hb_002471_310 |
0.0776711122 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001416_120 |
0.0790208181 |
- |
- |
caspase, putative [Ricinus communis] |
| 12 |
Hb_001496_150 |
0.0811366742 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 13 |
Hb_004693_040 |
0.0819804411 |
- |
- |
- |
| 14 |
Hb_014834_100 |
0.083555217 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 15 |
Hb_009158_010 |
0.0870370214 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 16 |
Hb_001557_030 |
0.0887366832 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 17 |
Hb_003152_010 |
0.0890290636 |
- |
- |
- |
| 18 |
Hb_007292_060 |
0.0893814763 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |
| 19 |
Hb_000796_110 |
0.0896308025 |
- |
- |
PREDICTED: probable pectinesterase 53 [Populus euphratica] |
| 20 |
Hb_002073_210 |
0.0900191609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |