| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
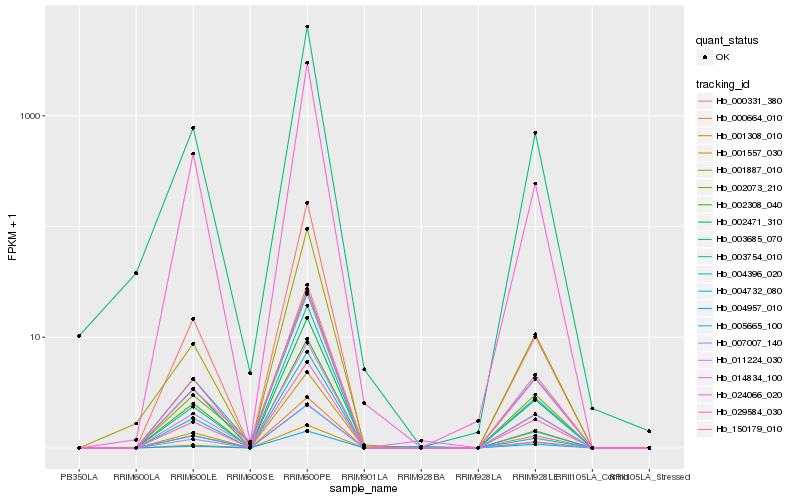
Hb_150179_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_21079 [Jatropha curcas] |
| 2 |
Hb_011224_030 |
0.04190314 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001557_030 |
0.0425342032 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 4 |
Hb_029584_030 |
0.0469926242 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 5 |
Hb_000664_010 |
0.0514767517 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 4-like [Jatropha curcas] |
| 6 |
Hb_007007_140 |
0.0576431303 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL6 [Jatropha curcas] |
| 7 |
Hb_003685_070 |
0.0628067895 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 8 |
Hb_002471_310 |
0.066425527 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002308_040 |
0.0670043482 |
- |
- |
methyladenine glycosylase family protein [Populus trichocarpa] |
| 10 |
Hb_001887_010 |
0.0694212621 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 11 |
Hb_024066_020 |
0.0694780112 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 12 |
Hb_004957_010 |
0.0715304386 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 13 |
Hb_003754_010 |
0.0719605556 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 14 |
Hb_014834_100 |
0.0727834406 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 15 |
Hb_004732_080 |
0.0776431815 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 16 |
Hb_004396_020 |
0.0793106383 |
- |
- |
SAUR-like auxin-responsive protein family [Theobroma cacao] |
| 17 |
Hb_002073_210 |
0.0797156044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005665_100 |
0.0806227001 |
- |
- |
PREDICTED: RING-H2 finger protein ATL63-like [Populus euphratica] |
| 19 |
Hb_001308_010 |
0.0863577582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000331_380 |
0.089572339 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |