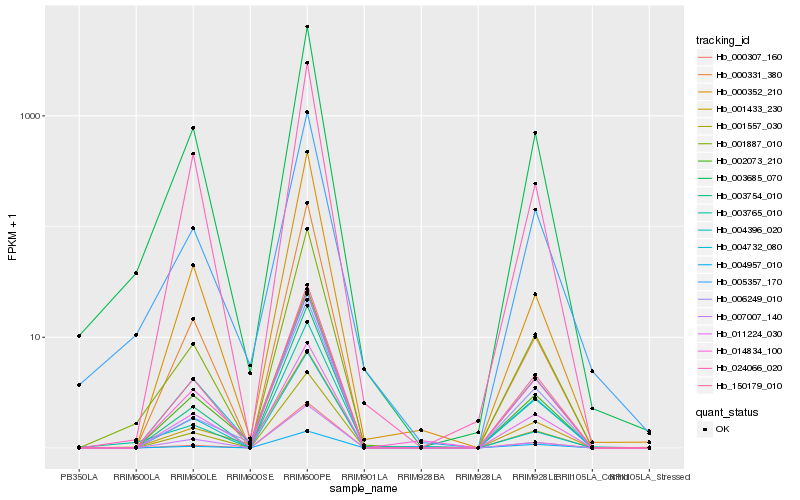
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001887_010 |
0.0 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 2 |
Hb_003685_070 |
0.0558048583 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 3 |
Hb_150179_010 |
0.0694212621 |
- |
- |
hypothetical protein JCGZ_21079 [Jatropha curcas] |
| 4 |
Hb_001557_030 |
0.0723951743 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 5 |
Hb_003765_010 |
0.078177032 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 6 |
Hb_005357_170 |
0.0784171698 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 7 |
Hb_002073_210 |
0.0787143778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000331_380 |
0.0847339551 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |
| 9 |
Hb_011224_030 |
0.0901215553 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007007_140 |
0.0902579326 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL6 [Jatropha curcas] |
| 11 |
Hb_003754_010 |
0.093667577 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 12 |
Hb_001433_230 |
0.0956913765 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 13 |
Hb_000307_160 |
0.0960335243 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 14 |
Hb_014834_100 |
0.09660531 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 15 |
Hb_000352_210 |
0.097264485 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 16 |
Hb_024066_020 |
0.0972683471 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 17 |
Hb_004396_020 |
0.0983867718 |
- |
- |
SAUR-like auxin-responsive protein family [Theobroma cacao] |
| 18 |
Hb_004957_010 |
0.1008361968 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 19 |
Hb_004732_080 |
0.1009483113 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 20 |
Hb_006249_010 |
0.1009494571 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |