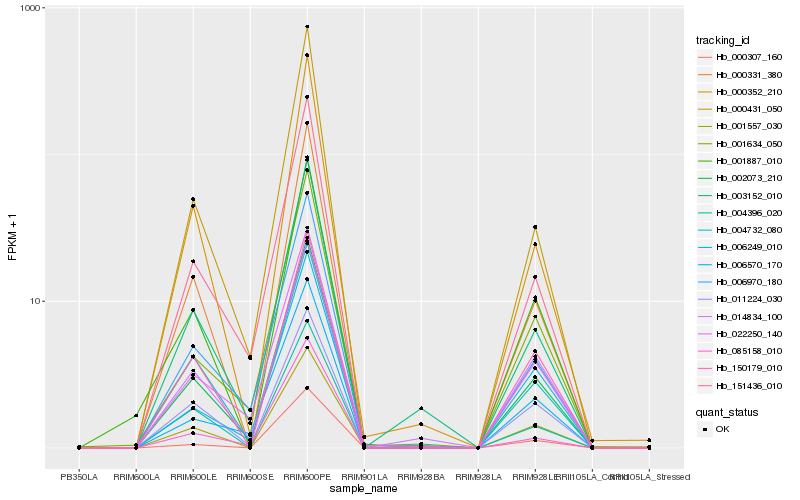
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002073_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_151436_010 |
0.0477768956 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 3 |
Hb_000331_380 |
0.0574634613 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |
| 4 |
Hb_006970_180 |
0.058412511 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 5 |
Hb_000431_050 |
0.0590168964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_022250_140 |
0.0639348542 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 7 |
Hb_000352_210 |
0.0675087105 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 8 |
Hb_003152_010 |
0.0707288097 |
- |
- |
- |
| 9 |
Hb_004732_080 |
0.072491841 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 10 |
Hb_001634_050 |
0.0726273608 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 11 |
Hb_006570_170 |
0.0762334402 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_085158_010 |
0.0768515172 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 13 |
Hb_006249_010 |
0.077181412 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 14 |
Hb_001557_030 |
0.0772200204 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 15 |
Hb_001887_010 |
0.0787143778 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 16 |
Hb_150179_010 |
0.0797156044 |
- |
- |
hypothetical protein JCGZ_21079 [Jatropha curcas] |
| 17 |
Hb_000307_160 |
0.0812562168 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 18 |
Hb_014834_100 |
0.082263997 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 19 |
Hb_011224_030 |
0.0831486724 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_004396_020 |
0.0848678593 |
- |
- |
SAUR-like auxin-responsive protein family [Theobroma cacao] |