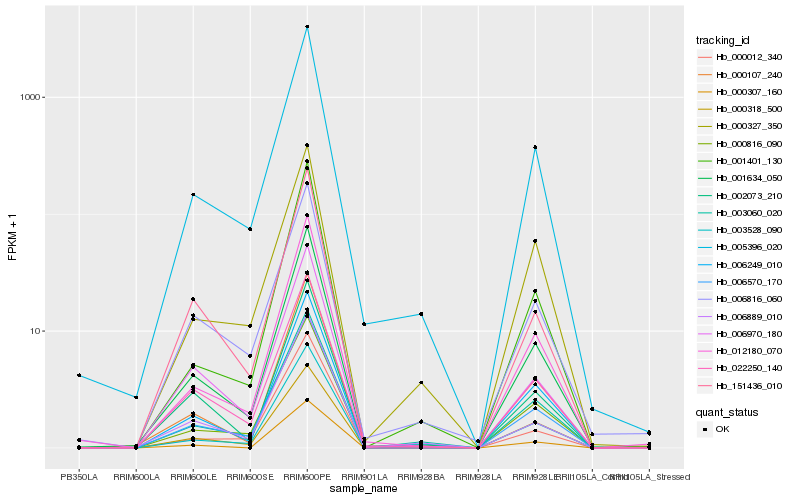
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006570_170 |
0.0 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_001634_050 |
0.0432138859 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 3 |
Hb_022250_140 |
0.0514175108 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 4 |
Hb_000816_090 |
0.0521025412 |
transcription factor |
TF Family: TRAF |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_005396_020 |
0.0587630023 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 6 |
Hb_006249_010 |
0.0589980004 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 7 |
Hb_012180_070 |
0.0668178156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003060_020 |
0.0674745656 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 9 |
Hb_151436_010 |
0.0717905099 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 10 |
Hb_006970_180 |
0.0744007988 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 11 |
Hb_000318_500 |
0.0745177637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002073_210 |
0.0762334402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006816_060 |
0.0791749111 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 14 |
Hb_000107_240 |
0.0800757559 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 15 |
Hb_001401_130 |
0.0858496165 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 16 |
Hb_003528_090 |
0.0858781275 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 17 |
Hb_000012_340 |
0.0901554217 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 18 |
Hb_006889_010 |
0.0910604663 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 19 |
Hb_000327_350 |
0.0913993677 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 20 |
Hb_000307_160 |
0.0930987575 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |