| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
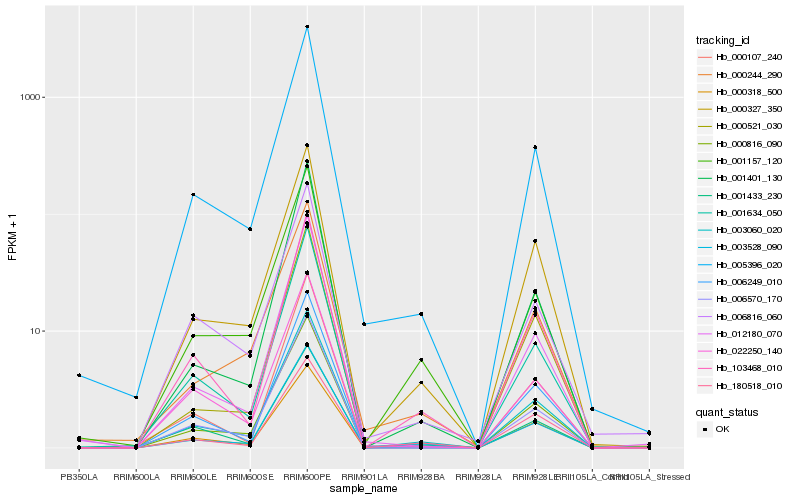
Hb_003060_020 |
0.0 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 2 |
Hb_003528_090 |
0.0381999 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 3 |
Hb_000816_090 |
0.0401616678 |
transcription factor |
TF Family: TRAF |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_000327_350 |
0.0514264382 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 5 |
Hb_005396_020 |
0.0557706312 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 6 |
Hb_103468_010 |
0.0565223647 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_001634_050 |
0.0664573806 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 8 |
Hb_001157_120 |
0.0672618372 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 9 |
Hb_006570_170 |
0.0674745656 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_006249_010 |
0.0763159073 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 11 |
Hb_022250_140 |
0.0798602154 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 12 |
Hb_000318_500 |
0.0813458244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000107_240 |
0.0828811802 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 14 |
Hb_012180_070 |
0.0834106269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006816_060 |
0.0842094047 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 16 |
Hb_001401_130 |
0.0853597273 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 17 |
Hb_180518_010 |
0.0891029561 |
- |
- |
PREDICTED: receptor-like protein 2 [Jatropha curcas] |
| 18 |
Hb_000244_290 |
0.0901413514 |
- |
- |
PREDICTED: UDP-glucuronate:xylan alpha-glucuronosyltransferase 1 [Jatropha curcas] |
| 19 |
Hb_001433_230 |
0.0901837758 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 20 |
Hb_000521_030 |
0.0912618943 |
- |
- |
PREDICTED: uncharacterized protein LOC105650350 [Jatropha curcas] |