| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
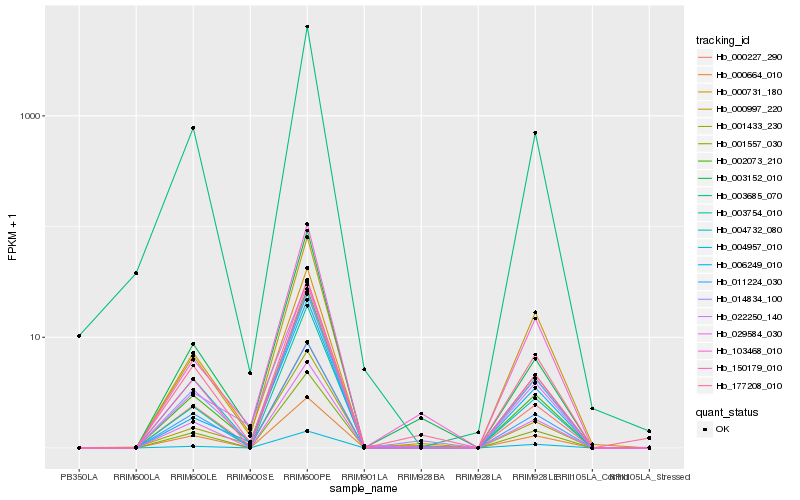
Hb_014834_100 |
0.0 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 2 |
Hb_001557_030 |
0.0535257858 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 3 |
Hb_103468_010 |
0.065579592 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_011224_030 |
0.0659186383 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001433_230 |
0.0686728178 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 6 |
Hb_150179_010 |
0.0727834406 |
- |
- |
hypothetical protein JCGZ_21079 [Jatropha curcas] |
| 7 |
Hb_002073_210 |
0.082263997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000731_180 |
0.083555217 |
- |
- |
hypothetical protein POPTR_0004s01820g [Populus trichocarpa] |
| 9 |
Hb_177208_010 |
0.0840519082 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor, partial [Jatropha sp. CONN 198700232] |
| 10 |
Hb_022250_140 |
0.0841630964 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 11 |
Hb_004732_080 |
0.0851746206 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 12 |
Hb_000997_220 |
0.085410159 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 13 |
Hb_000227_290 |
0.0871443732 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 14 |
Hb_003152_010 |
0.0885220341 |
- |
- |
- |
| 15 |
Hb_029584_030 |
0.0895366118 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 16 |
Hb_003754_010 |
0.090195177 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 17 |
Hb_004957_010 |
0.0907504434 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 18 |
Hb_003685_070 |
0.0933863385 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 19 |
Hb_006249_010 |
0.0937503465 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 20 |
Hb_000664_010 |
0.094626606 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 4-like [Jatropha curcas] |