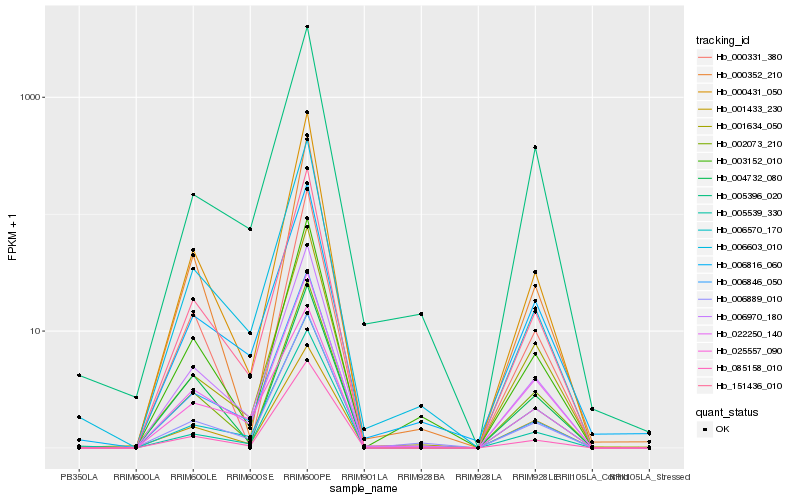
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006970_180 |
0.0 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 2 |
Hb_151436_010 |
0.0253851344 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 3 |
Hb_022250_140 |
0.0543142749 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 4 |
Hb_085158_010 |
0.0544977348 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 5 |
Hb_000431_050 |
0.0570233935 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006603_010 |
0.0577554555 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 7 |
Hb_002073_210 |
0.058412511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006889_010 |
0.0631885453 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 9 |
Hb_003152_010 |
0.0694890089 |
- |
- |
- |
| 10 |
Hb_006570_170 |
0.0744007988 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_006846_050 |
0.0757645372 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 12 |
Hb_001634_050 |
0.0776145947 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 13 |
Hb_000352_210 |
0.0807378312 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 14 |
Hb_006816_060 |
0.0825165865 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 15 |
Hb_000331_380 |
0.0858347503 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |
| 16 |
Hb_025557_090 |
0.0860536621 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |
| 17 |
Hb_005539_330 |
0.0908191069 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 18 |
Hb_004732_080 |
0.0925045881 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 19 |
Hb_005396_020 |
0.0925396633 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 20 |
Hb_001433_230 |
0.0931818547 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |