| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
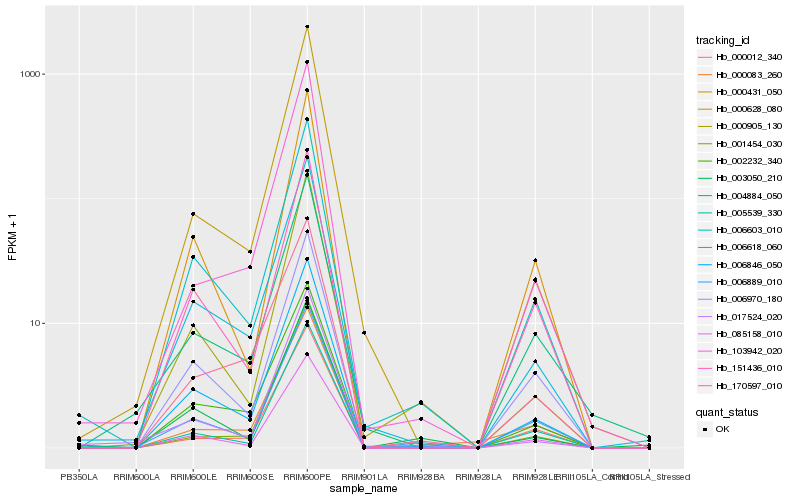
Hb_006846_050 |
0.0 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 2 |
Hb_006618_060 |
0.0535267439 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 3 |
Hb_006603_010 |
0.0542178121 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 4 |
Hb_085158_010 |
0.0584449124 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 5 |
Hb_002232_340 |
0.0627392599 |
- |
- |
Sodium transporter hkt1-like protein, putative [Theobroma cacao] |
| 6 |
Hb_006970_180 |
0.0757645372 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 7 |
Hb_151436_010 |
0.0784820256 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 8 |
Hb_006889_010 |
0.0805204667 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 9 |
Hb_000431_050 |
0.0810218791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000905_130 |
0.084249403 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100 [Jatropha curcas] |
| 11 |
Hb_000628_080 |
0.086125003 |
- |
- |
hypothetical protein JCGZ_04283 [Jatropha curcas] |
| 12 |
Hb_005539_330 |
0.0907397172 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 13 |
Hb_170597_010 |
0.0921270434 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 14 |
Hb_003050_210 |
0.0977873792 |
- |
- |
PREDICTED: cytochrome b561 domain-containing protein At4g18260-like [Populus euphratica] |
| 15 |
Hb_001454_030 |
0.1002091432 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 3 [Jatropha curcas] |
| 16 |
Hb_000012_340 |
0.1008133099 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 17 |
Hb_017524_020 |
0.1011790562 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 18 |
Hb_103942_020 |
0.1026918769 |
- |
- |
unknown [Zea mays] |
| 19 |
Hb_000083_260 |
0.1039451872 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004884_050 |
0.1042823088 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |