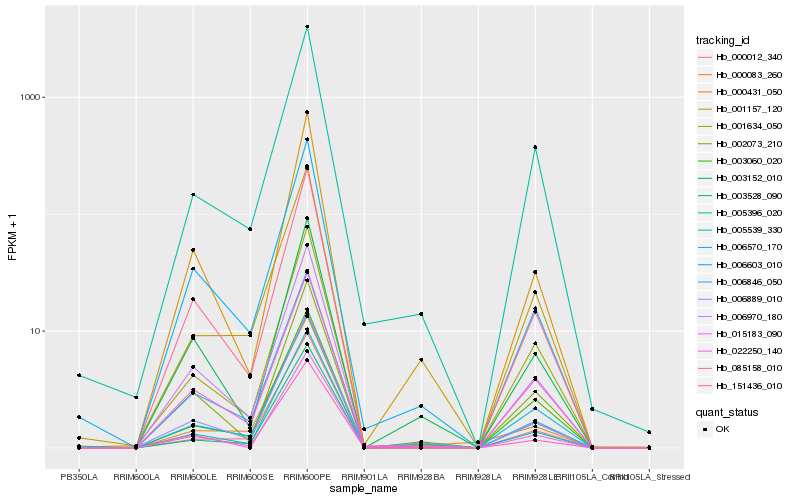
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006889_010 |
0.0 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 2 |
Hb_006970_180 |
0.0631885453 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 3 |
Hb_003152_010 |
0.0654234963 |
- |
- |
- |
| 4 |
Hb_005539_330 |
0.0698131047 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 5 |
Hb_151436_010 |
0.0699224245 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 6 |
Hb_085158_010 |
0.0701813999 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 7 |
Hb_006603_010 |
0.070523777 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 8 |
Hb_000012_340 |
0.0778774082 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 9 |
Hb_000431_050 |
0.0788119854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006846_050 |
0.0805204667 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 11 |
Hb_001634_050 |
0.0835976839 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 12 |
Hb_005396_020 |
0.0887230264 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 13 |
Hb_015183_090 |
0.0893674035 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 14 |
Hb_022250_140 |
0.0897402792 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 15 |
Hb_006570_170 |
0.0910604663 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_001157_120 |
0.092120721 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 17 |
Hb_002073_210 |
0.0926260386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003060_020 |
0.0948902332 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_000083_260 |
0.0951067912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003528_090 |
0.0959048327 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |