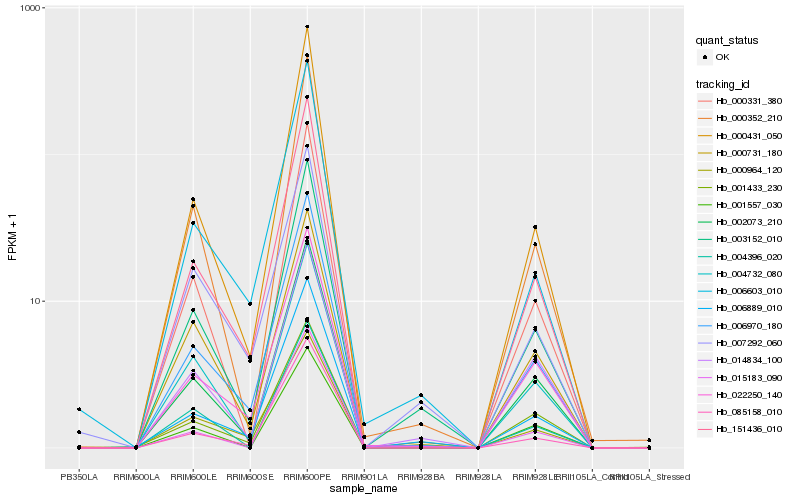
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003152_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_006889_010 |
0.0654234963 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 3 |
Hb_000352_210 |
0.0689197361 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 4 |
Hb_006970_180 |
0.0694890089 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 5 |
Hb_002073_210 |
0.0707288097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_151436_010 |
0.0733535606 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 7 |
Hb_000431_050 |
0.0752964131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004732_080 |
0.0770672915 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 9 |
Hb_015183_090 |
0.0793428898 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 10 |
Hb_000331_380 |
0.0799277325 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |
| 11 |
Hb_006603_010 |
0.0866855864 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 12 |
Hb_085158_010 |
0.0877036412 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 13 |
Hb_014834_100 |
0.0885220341 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 14 |
Hb_000731_180 |
0.0890290636 |
- |
- |
hypothetical protein POPTR_0004s01820g [Populus trichocarpa] |
| 15 |
Hb_001557_030 |
0.0898020274 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 16 |
Hb_022250_140 |
0.0919495572 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 17 |
Hb_001433_230 |
0.0948029563 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 18 |
Hb_007292_060 |
0.0959566703 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |
| 19 |
Hb_000964_120 |
0.0962992439 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 20 |
Hb_004396_020 |
0.0974760819 |
- |
- |
SAUR-like auxin-responsive protein family [Theobroma cacao] |