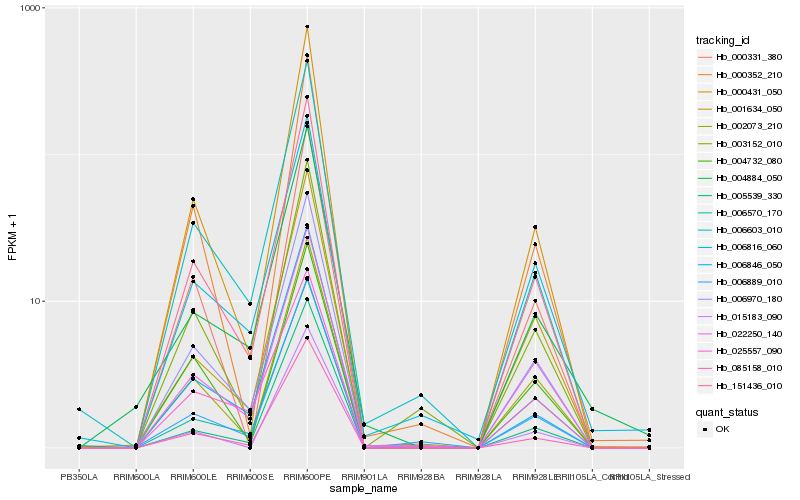
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_151436_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 2 |
Hb_006970_180 |
0.0253851344 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 3 |
Hb_000431_050 |
0.0441278266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_085158_010 |
0.0442691842 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 5 |
Hb_002073_210 |
0.0477768956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_022250_140 |
0.0636986638 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 7 |
Hb_006603_010 |
0.0698049268 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 8 |
Hb_006889_010 |
0.0699224245 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 9 |
Hb_006570_170 |
0.0717905099 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_003152_010 |
0.0733535606 |
- |
- |
- |
| 11 |
Hb_000331_380 |
0.0758751398 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |
| 12 |
Hb_000352_210 |
0.078243256 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 13 |
Hb_006846_050 |
0.0784820256 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 14 |
Hb_001634_050 |
0.0790848974 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 15 |
Hb_025557_090 |
0.0907997814 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |
| 16 |
Hb_005539_330 |
0.091192371 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 17 |
Hb_004732_080 |
0.0916239752 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 18 |
Hb_006816_060 |
0.0939385052 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 19 |
Hb_015183_090 |
0.0986253835 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 20 |
Hb_004884_050 |
0.0994293815 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |