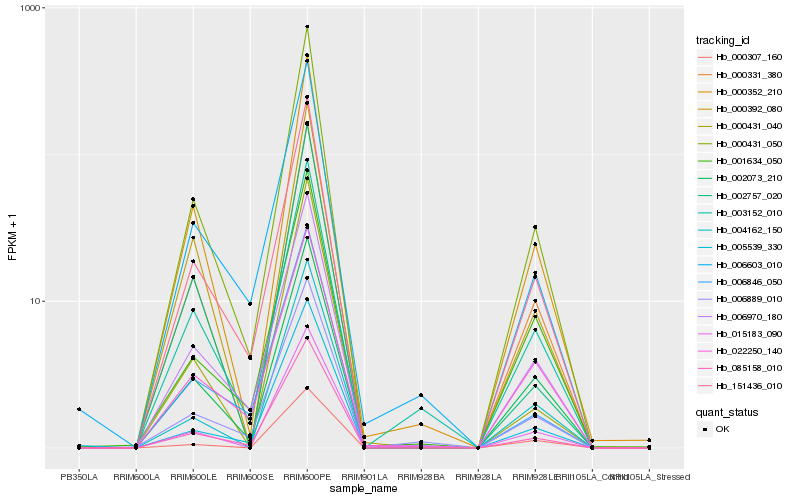
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000431_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_085158_010 |
0.0290338943 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 3 |
Hb_151436_010 |
0.0441278266 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 4 |
Hb_000331_380 |
0.0563498834 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |
| 5 |
Hb_006970_180 |
0.0570233935 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 6 |
Hb_002073_210 |
0.0590168964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000352_210 |
0.0592784675 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 8 |
Hb_003152_010 |
0.0752964131 |
- |
- |
- |
| 9 |
Hb_004162_150 |
0.0766707875 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 10 |
Hb_015183_090 |
0.0768687401 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 11 |
Hb_006889_010 |
0.0788119854 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 12 |
Hb_006846_050 |
0.0810218791 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 13 |
Hb_006603_010 |
0.0810920176 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 14 |
Hb_000392_080 |
0.0824447699 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |
| 15 |
Hb_005539_330 |
0.0835247376 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 16 |
Hb_000431_040 |
0.0906663313 |
- |
- |
PREDICTED: uncharacterized protein LOC105635551 [Jatropha curcas] |
| 17 |
Hb_000307_160 |
0.0934238893 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 18 |
Hb_001634_050 |
0.0957214465 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 19 |
Hb_002757_020 |
0.096692843 |
- |
- |
- |
| 20 |
Hb_022250_140 |
0.0987192395 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |