| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
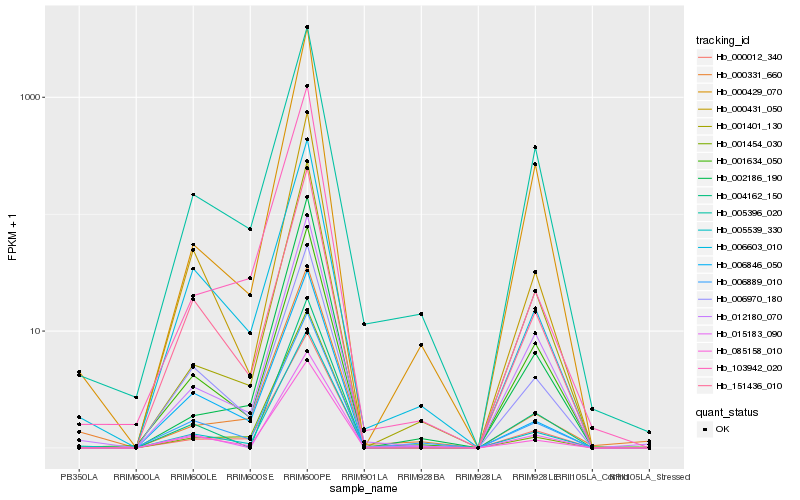
Hb_005539_330 |
0.0 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 2 |
Hb_006889_010 |
0.0698131047 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 3 |
Hb_000012_340 |
0.0710998472 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 4 |
Hb_085158_010 |
0.0714491938 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 5 |
Hb_000429_070 |
0.0816458285 |
- |
- |
hypothetical protein POPTR_0006s05480g [Populus trichocarpa] |
| 6 |
Hb_000431_050 |
0.0835247376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006603_010 |
0.0856191802 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 8 |
Hb_001401_130 |
0.0895955428 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 9 |
Hb_004162_150 |
0.0902639018 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 10 |
Hb_006846_050 |
0.0907397172 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 11 |
Hb_006970_180 |
0.0908191069 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 12 |
Hb_151436_010 |
0.091192371 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 13 |
Hb_015183_090 |
0.0922659069 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 14 |
Hb_001634_050 |
0.09237466 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 15 |
Hb_012180_070 |
0.0934863618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002186_190 |
0.0947858723 |
- |
- |
- |
| 17 |
Hb_000331_660 |
0.0974217005 |
- |
- |
PREDICTED: vignain [Jatropha curcas] |
| 18 |
Hb_001454_030 |
0.0982052779 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 3 [Jatropha curcas] |
| 19 |
Hb_103942_020 |
0.0983371594 |
- |
- |
unknown [Zea mays] |
| 20 |
Hb_005396_020 |
0.0988520036 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |