| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
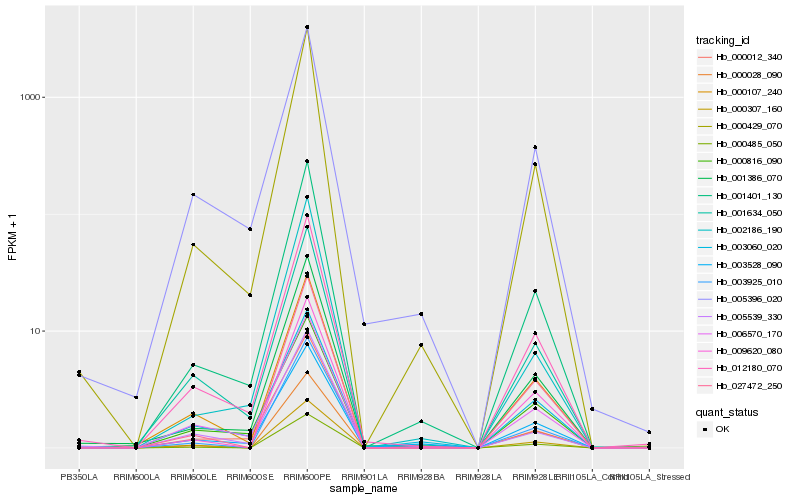
Hb_001401_130 |
0.0 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 2 |
Hb_000429_070 |
0.0298017944 |
- |
- |
hypothetical protein POPTR_0006s05480g [Populus trichocarpa] |
| 3 |
Hb_001386_070 |
0.0560094821 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_012180_070 |
0.058080684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003528_090 |
0.0646715192 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 6 |
Hb_002186_190 |
0.0682376647 |
- |
- |
- |
| 7 |
Hb_000012_340 |
0.0690484566 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 8 |
Hb_000107_240 |
0.0710598895 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 9 |
Hb_000485_050 |
0.0713837256 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 10 |
Hb_001634_050 |
0.0739873151 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 11 |
Hb_003925_010 |
0.0788939014 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 12 |
Hb_027472_250 |
0.0789772393 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 13 |
Hb_005396_020 |
0.0793730835 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 14 |
Hb_000816_090 |
0.0850040857 |
transcription factor |
TF Family: TRAF |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_003060_020 |
0.0853597273 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 16 |
Hb_009620_080 |
0.0854941893 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 17 |
Hb_006570_170 |
0.0858496165 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000028_090 |
0.0871386917 |
- |
- |
PREDICTED: uncharacterized protein LOC105135810 [Populus euphratica] |
| 19 |
Hb_000307_160 |
0.0889340672 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 20 |
Hb_005539_330 |
0.0895955428 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |