| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
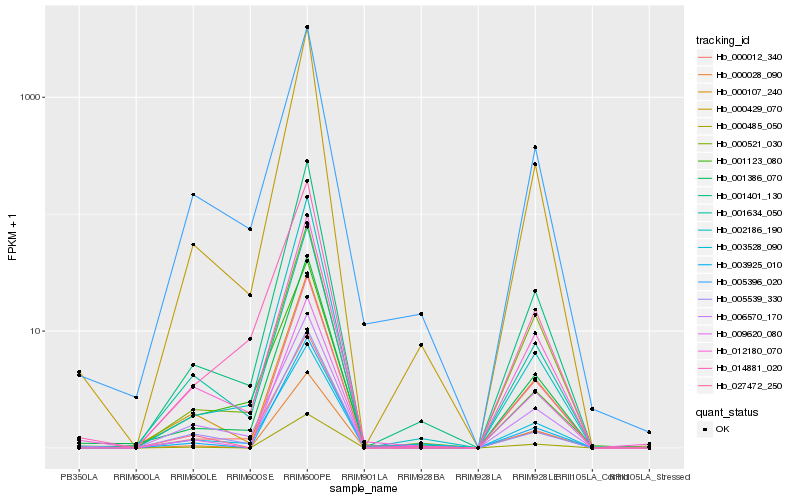
Hb_001386_070 |
0.0 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_000429_070 |
0.0512378913 |
- |
- |
hypothetical protein POPTR_0006s05480g [Populus trichocarpa] |
| 3 |
Hb_001401_130 |
0.0560094821 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 4 |
Hb_012180_070 |
0.0567377843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002186_190 |
0.0816550322 |
- |
- |
- |
| 6 |
Hb_003925_010 |
0.0840204352 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 7 |
Hb_000485_050 |
0.0858485681 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 8 |
Hb_001634_050 |
0.086135736 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 9 |
Hb_005396_020 |
0.0862022708 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 10 |
Hb_014881_020 |
0.0868528393 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 11 |
Hb_000107_240 |
0.0869467514 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 12 |
Hb_027472_250 |
0.0886047708 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 13 |
Hb_000012_340 |
0.0925976361 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 14 |
Hb_009620_080 |
0.0937482507 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 15 |
Hb_000028_090 |
0.097054394 |
- |
- |
PREDICTED: uncharacterized protein LOC105135810 [Populus euphratica] |
| 16 |
Hb_003528_090 |
0.0972476644 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 17 |
Hb_001123_080 |
0.098625099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005539_330 |
0.1010960036 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 19 |
Hb_006570_170 |
0.1022054267 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000521_030 |
0.1026445413 |
- |
- |
PREDICTED: uncharacterized protein LOC105650350 [Jatropha curcas] |