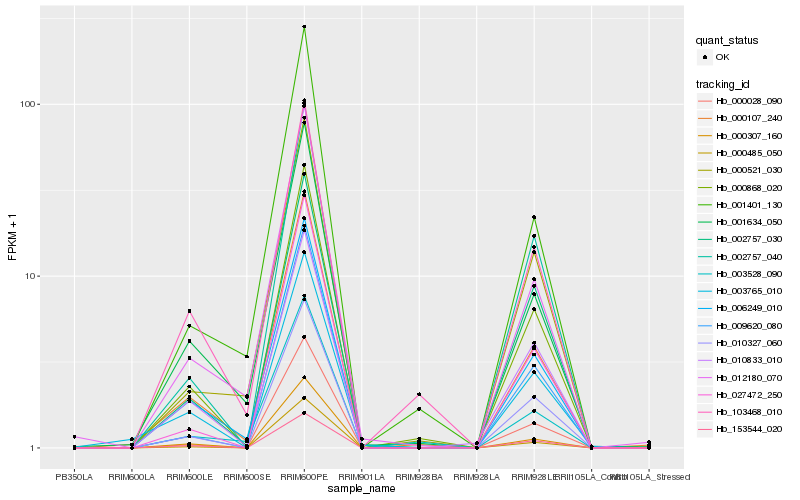
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000868_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_010327_060 |
0.0486865984 |
- |
- |
hypothetical protein B456_001G246300 [Gossypium raimondii] |
| 3 |
Hb_000028_090 |
0.0593977807 |
- |
- |
PREDICTED: uncharacterized protein LOC105135810 [Populus euphratica] |
| 4 |
Hb_000107_240 |
0.063154691 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 5 |
Hb_002757_040 |
0.0632467124 |
- |
- |
- |
| 6 |
Hb_000307_160 |
0.0671652631 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 7 |
Hb_006249_010 |
0.0684694136 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 8 |
Hb_153544_020 |
0.0693645871 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP12 [Jatropha curcas] |
| 9 |
Hb_103468_010 |
0.0708055072 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_009620_080 |
0.0714910244 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 11 |
Hb_000485_050 |
0.0720550163 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 12 |
Hb_027472_250 |
0.0732276312 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 13 |
Hb_010833_010 |
0.0839968755 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 14 |
Hb_002757_030 |
0.0852856404 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000521_030 |
0.0907148708 |
- |
- |
PREDICTED: uncharacterized protein LOC105650350 [Jatropha curcas] |
| 16 |
Hb_001634_050 |
0.0910573078 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 17 |
Hb_003765_010 |
0.0912062594 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 18 |
Hb_012180_070 |
0.0945273651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003528_090 |
0.0948302783 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 20 |
Hb_001401_130 |
0.0951073671 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |