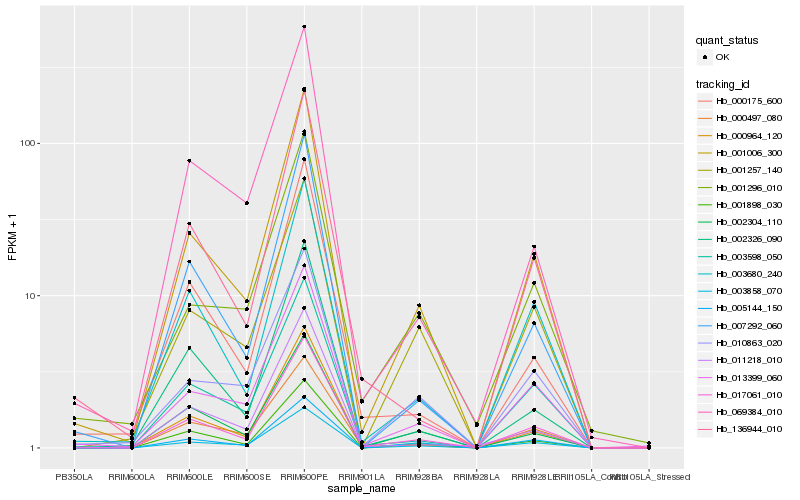
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001006_300 |
0.0 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 2 |
Hb_017061_010 |
0.0471565634 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 3 |
Hb_003858_070 |
0.0537970033 |
- |
- |
PREDICTED: uncharacterized protein LOC105647879 [Jatropha curcas] |
| 4 |
Hb_001898_030 |
0.0712313979 |
- |
- |
protein kinase-coding resistance protein [Nicotiana repanda] |
| 5 |
Hb_013399_060 |
0.0724905163 |
- |
- |
PREDICTED: uncharacterized protein LOC105133408 [Populus euphratica] |
| 6 |
Hb_000964_120 |
0.0775298142 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 7 |
Hb_005144_150 |
0.0800326802 |
- |
- |
hypothetical protein VITISV_013572 [Vitis vinifera] |
| 8 |
Hb_007292_060 |
0.0908275984 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |
| 9 |
Hb_010863_020 |
0.0966731632 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_003598_050 |
0.0994016326 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 11 |
Hb_002304_110 |
0.1003448528 |
- |
- |
hypothetical protein MIMGU_mgv1a023246mg [Erythranthe guttata] |
| 12 |
Hb_003680_240 |
0.100431318 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 13 |
Hb_000497_080 |
0.105345233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011218_010 |
0.1059308659 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 15 |
Hb_002326_090 |
0.1083670565 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_001257_140 |
0.1102367805 |
- |
- |
Gb:AAF02129.1, putative [Theobroma cacao] |
| 17 |
Hb_001296_010 |
0.1111724441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_069384_010 |
0.1114610672 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_136944_010 |
0.1128134517 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 20 |
Hb_000175_600 |
0.1134205729 |
- |
- |
PREDICTED: TATA-binding protein-associated factor 2N isoform X2 [Jatropha curcas] |