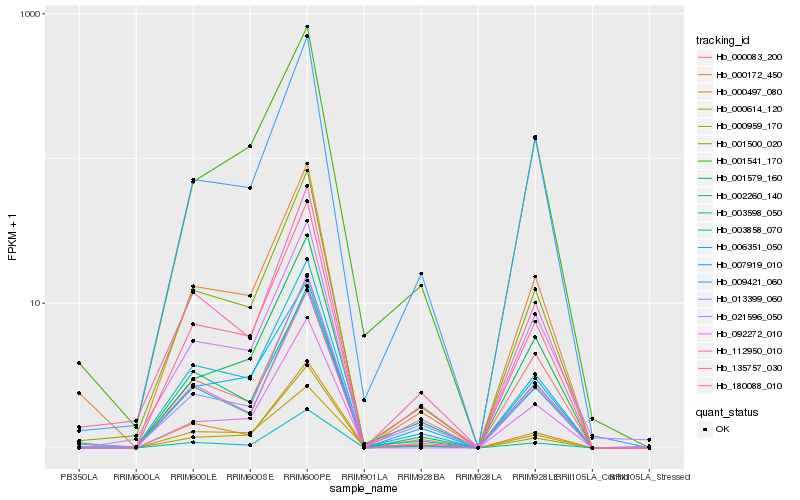
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_180088_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_006351_050 |
0.0420521352 |
- |
- |
PREDICTED: uncharacterized protein LOC105133646 [Populus euphratica] |
| 3 |
Hb_001500_020 |
0.0500472881 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_007919_010 |
0.0601499067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_112950_010 |
0.0703942705 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 6 |
Hb_009421_060 |
0.0724926081 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 7 |
Hb_003598_050 |
0.0755564466 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 8 |
Hb_003858_070 |
0.0796608474 |
- |
- |
PREDICTED: uncharacterized protein LOC105647879 [Jatropha curcas] |
| 9 |
Hb_001579_160 |
0.080121085 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 10 |
Hb_000497_080 |
0.0838885334 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000172_450 |
0.0852580209 |
- |
- |
Uncharacterized protein TCM_001203 [Theobroma cacao] |
| 12 |
Hb_013399_060 |
0.0867978354 |
- |
- |
PREDICTED: uncharacterized protein LOC105133408 [Populus euphratica] |
| 13 |
Hb_002260_140 |
0.0887420809 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_135757_030 |
0.0895908819 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 15 |
Hb_021596_050 |
0.0906070637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000083_200 |
0.0982963418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001541_170 |
0.0989185223 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 18 |
Hb_000959_170 |
0.0993577591 |
- |
- |
Cell division protein kinase 7, putative [Ricinus communis] |
| 19 |
Hb_000614_120 |
0.1029639099 |
- |
- |
hypothetical protein JCGZ_17923 [Jatropha curcas] |
| 20 |
Hb_092272_010 |
0.1036443966 |
- |
- |
- |