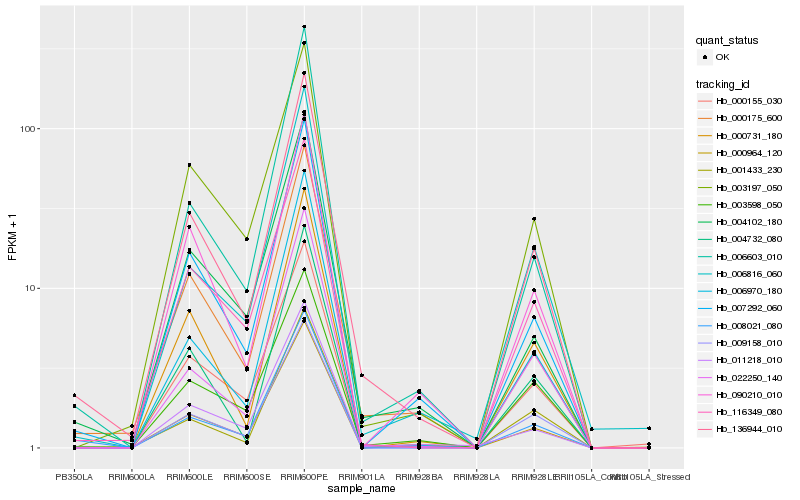
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_136944_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 2 |
Hb_000175_600 |
0.0753721274 |
- |
- |
PREDICTED: TATA-binding protein-associated factor 2N isoform X2 [Jatropha curcas] |
| 3 |
Hb_007292_060 |
0.0830232444 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |
| 4 |
Hb_116349_080 |
0.0865218787 |
- |
- |
Nectarin-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000964_120 |
0.0903610709 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 6 |
Hb_004102_180 |
0.0906074665 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 7 |
Hb_000731_180 |
0.0923321039 |
- |
- |
hypothetical protein POPTR_0004s01820g [Populus trichocarpa] |
| 8 |
Hb_003197_050 |
0.0938609059 |
- |
- |
hypothetical protein JCGZ_04279 [Jatropha curcas] |
| 9 |
Hb_011218_010 |
0.0960799662 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 10 |
Hb_009158_010 |
0.0974851304 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 11 |
Hb_008021_080 |
0.0976641136 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Nicotiana sylvestris] |
| 12 |
Hb_001433_230 |
0.0980932361 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 13 |
Hb_090210_010 |
0.0998497855 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 14 |
Hb_022250_140 |
0.0998786754 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 15 |
Hb_006970_180 |
0.1010337094 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 16 |
Hb_000155_030 |
0.1015340499 |
- |
- |
PREDICTED: uncharacterized protein LOC105649591 [Jatropha curcas] |
| 17 |
Hb_003598_050 |
0.1016471922 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 18 |
Hb_006603_010 |
0.1026044398 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 19 |
Hb_004732_080 |
0.1030567505 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 20 |
Hb_006816_060 |
0.1042104846 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |