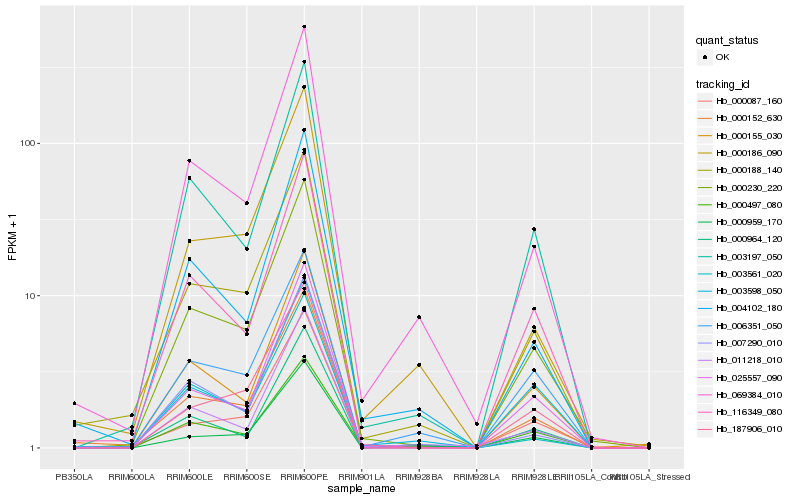
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_220 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003197_050 |
0.0709577417 |
- |
- |
hypothetical protein JCGZ_04279 [Jatropha curcas] |
| 3 |
Hb_000152_630 |
0.0737213701 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Jatropha curcas] |
| 4 |
Hb_116349_080 |
0.0749375148 |
- |
- |
Nectarin-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000188_140 |
0.0758962796 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 6 |
Hb_000155_030 |
0.0834929024 |
- |
- |
PREDICTED: uncharacterized protein LOC105649591 [Jatropha curcas] |
| 7 |
Hb_025557_090 |
0.0848459213 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |
| 8 |
Hb_069384_010 |
0.0857352823 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000087_160 |
0.0894375304 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_187906_010 |
0.0921631221 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA2-like [Populus euphratica] |
| 11 |
Hb_000497_080 |
0.093805183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006351_050 |
0.0977552564 |
- |
- |
PREDICTED: uncharacterized protein LOC105133646 [Populus euphratica] |
| 13 |
Hb_004102_180 |
0.1000542911 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 14 |
Hb_011218_010 |
0.100933803 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 15 |
Hb_007290_010 |
0.1026322651 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000959_170 |
0.1044742869 |
- |
- |
Cell division protein kinase 7, putative [Ricinus communis] |
| 17 |
Hb_000186_090 |
0.1073901866 |
- |
- |
ACC oxidase 1 [Hevea brasiliensis] |
| 18 |
Hb_000964_120 |
0.1089827026 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 19 |
Hb_003561_020 |
0.11012929 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 20 |
Hb_003598_050 |
0.1115769854 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |