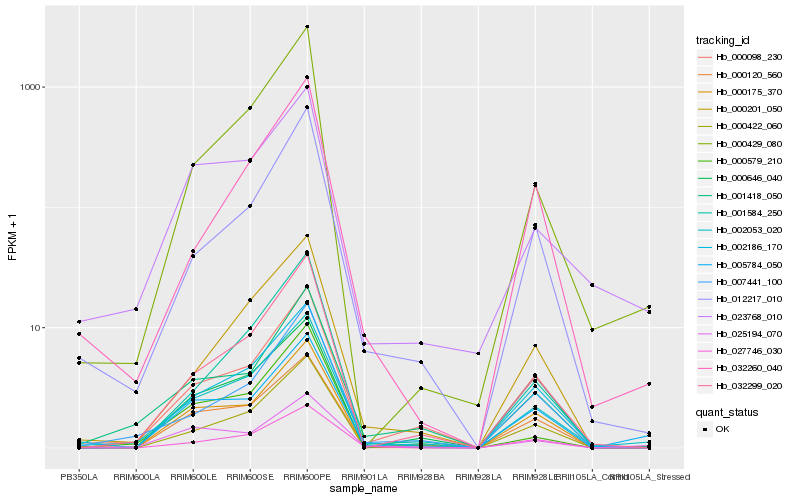
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002053_020 |
0.0 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 2 |
Hb_002186_170 |
0.1002259441 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 3 |
Hb_000175_370 |
0.1100474243 |
- |
- |
hypothetical protein EUGRSUZ_F03862 [Eucalyptus grandis] |
| 4 |
Hb_032299_020 |
0.1114982501 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 5 |
Hb_005784_050 |
0.1133366949 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_000429_080 |
0.1142878769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_023768_010 |
0.1162858508 |
- |
- |
- |
| 8 |
Hb_000201_050 |
0.1163950674 |
- |
- |
- |
| 9 |
Hb_000098_230 |
0.1170114353 |
- |
- |
Potassium channel AKT2/3, putative [Ricinus communis] |
| 10 |
Hb_000120_560 |
0.1180309029 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_000646_040 |
0.1186591393 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 12 |
Hb_027746_030 |
0.1202666267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012217_010 |
0.1216599455 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 14 |
Hb_007441_100 |
0.1273271248 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 12 [Populus euphratica] |
| 15 |
Hb_001418_050 |
0.1276560342 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 16 |
Hb_000422_060 |
0.1314227569 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 17 |
Hb_025194_070 |
0.1346652711 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4-like [Jatropha curcas] |
| 18 |
Hb_032260_040 |
0.1362097725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000579_210 |
0.1365416286 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 20 |
Hb_001584_250 |
0.1380221013 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |