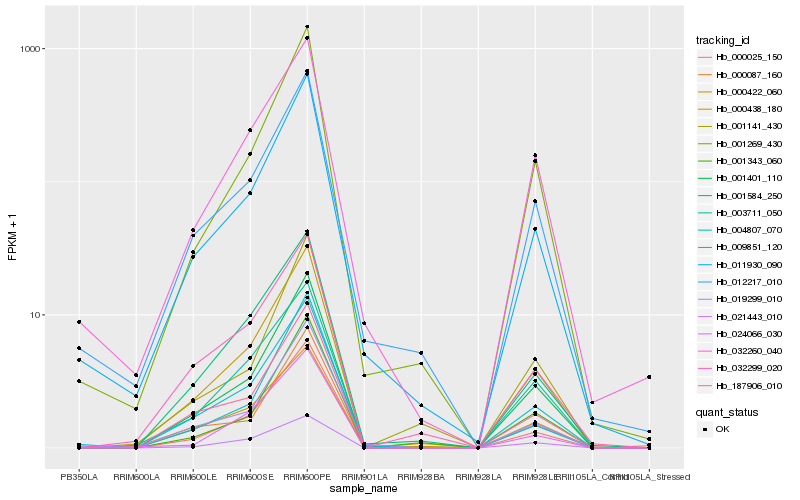
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000438_180 |
0.0 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_001401_110 |
0.043688531 |
- |
- |
hypothetical protein JCGZ_04272 [Jatropha curcas] |
| 3 |
Hb_187906_010 |
0.0717527743 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA2-like [Populus euphratica] |
| 4 |
Hb_001269_430 |
0.0743119776 |
- |
- |
hypothetical protein JCGZ_04286 [Jatropha curcas] |
| 5 |
Hb_003711_050 |
0.0746863407 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 6 |
Hb_000025_150 |
0.0774602737 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 7 |
Hb_021443_010 |
0.0779406306 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 8 |
Hb_001584_250 |
0.0782668044 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 9 |
Hb_004807_070 |
0.0820975395 |
- |
- |
hypothetical protein POPTR_0004s17330g, partial [Populus trichocarpa] |
| 10 |
Hb_011930_090 |
0.082487106 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 11 |
Hb_032299_020 |
0.0848749242 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 12 |
Hb_012217_010 |
0.0913149035 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 13 |
Hb_032260_040 |
0.0951790954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000087_160 |
0.0961849685 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_019299_010 |
0.0965456295 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000422_060 |
0.099989595 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 17 |
Hb_009851_120 |
0.1043006626 |
- |
- |
PREDICTED: RING-H2 zinc finger protein RHA4a [Jatropha curcas] |
| 18 |
Hb_001343_060 |
0.1048400259 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 19 |
Hb_001141_430 |
0.1061594667 |
- |
- |
hypothetical protein POPTR_0008s15160g [Populus trichocarpa] |
| 20 |
Hb_024066_030 |
0.1063061776 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |