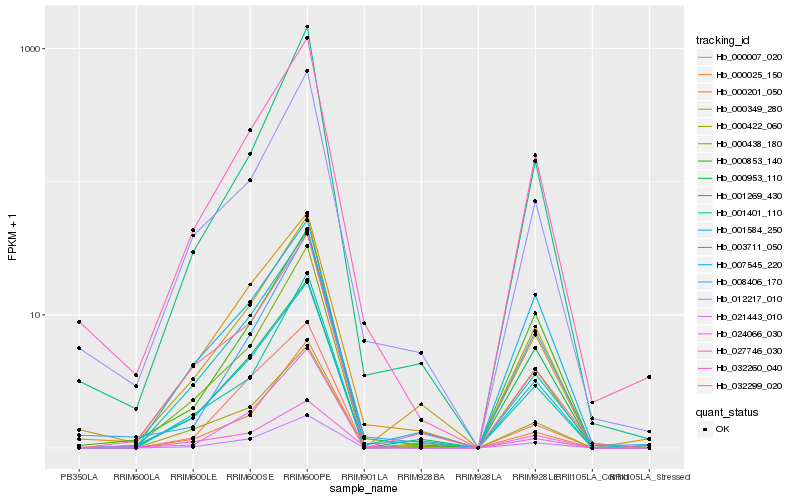
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021443_010 |
0.0 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 2 |
Hb_003711_050 |
0.0487909812 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 3 |
Hb_000438_180 |
0.0779406306 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_032260_040 |
0.0859955179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001584_250 |
0.0948911604 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 6 |
Hb_001401_110 |
0.0953648537 |
- |
- |
hypothetical protein JCGZ_04272 [Jatropha curcas] |
| 7 |
Hb_000201_050 |
0.1017698035 |
- |
- |
- |
| 8 |
Hb_000007_020 |
0.1040407751 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 9 |
Hb_000853_140 |
0.1075306344 |
- |
- |
hypothetical protein POPTR_0014s12080g [Populus trichocarpa] |
| 10 |
Hb_024066_030 |
0.1107622043 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 11 |
Hb_001269_430 |
0.1138205545 |
- |
- |
hypothetical protein JCGZ_04286 [Jatropha curcas] |
| 12 |
Hb_000349_280 |
0.1138715796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000422_060 |
0.114208031 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 14 |
Hb_000025_150 |
0.1192184719 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 15 |
Hb_008406_170 |
0.1218506757 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_032299_020 |
0.1243658879 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 17 |
Hb_027746_030 |
0.1243764287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000953_110 |
0.1245943563 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 19 |
Hb_007545_220 |
0.1278374382 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 20 |
Hb_012217_010 |
0.128608762 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |