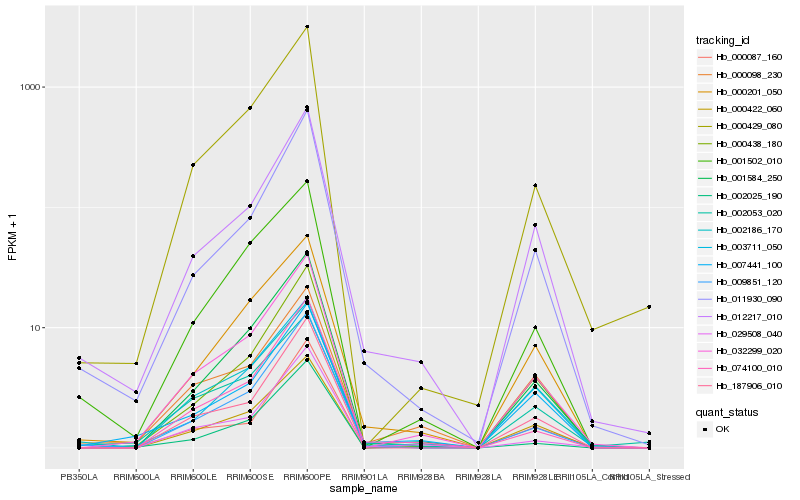
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032299_020 |
0.0 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 2 |
Hb_001584_250 |
0.069586983 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 3 |
Hb_000422_060 |
0.0706015057 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 4 |
Hb_000429_080 |
0.0773613324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000438_180 |
0.0848749242 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_187906_010 |
0.0897002746 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA2-like [Populus euphratica] |
| 7 |
Hb_012217_010 |
0.0976120428 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 8 |
Hb_000201_050 |
0.1027131999 |
- |
- |
- |
| 9 |
Hb_002186_170 |
0.1069552388 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 10 |
Hb_001502_010 |
0.1079581552 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 11 |
Hb_003711_050 |
0.1082003514 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 12 |
Hb_007441_100 |
0.1092991143 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 12 [Populus euphratica] |
| 13 |
Hb_029508_040 |
0.1094487344 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 14 |
Hb_011930_090 |
0.1098924123 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 15 |
Hb_009851_120 |
0.1104847071 |
- |
- |
PREDICTED: RING-H2 zinc finger protein RHA4a [Jatropha curcas] |
| 16 |
Hb_000087_160 |
0.1105253181 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_002053_020 |
0.1114982501 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 18 |
Hb_002025_190 |
0.1115824389 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 19 |
Hb_074100_010 |
0.1133094331 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 20 |
Hb_000098_230 |
0.1133338287 |
- |
- |
Potassium channel AKT2/3, putative [Ricinus communis] |