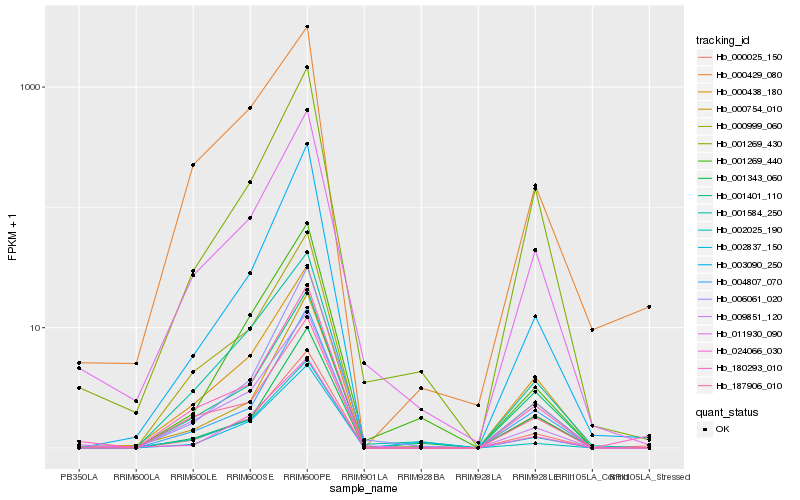
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_150 |
0.0 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 2 |
Hb_000438_180 |
0.0774602737 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_180293_010 |
0.0817260938 |
- |
- |
hypothetical protein POPTR_0008s19020g [Populus trichocarpa] |
| 4 |
Hb_001401_110 |
0.0889062893 |
- |
- |
hypothetical protein JCGZ_04272 [Jatropha curcas] |
| 5 |
Hb_001584_250 |
0.0923774254 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 6 |
Hb_001269_430 |
0.094370056 |
- |
- |
hypothetical protein JCGZ_04286 [Jatropha curcas] |
| 7 |
Hb_004807_070 |
0.0945335609 |
- |
- |
hypothetical protein POPTR_0004s17330g, partial [Populus trichocarpa] |
| 8 |
Hb_006061_020 |
0.0972044961 |
transcription factor |
TF Family: bZIP |
bZIP family protein [Populus trichocarpa] |
| 9 |
Hb_011930_090 |
0.0984252257 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 10 |
Hb_001269_440 |
0.1002590768 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 11 |
Hb_187906_010 |
0.1009149588 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA2-like [Populus euphratica] |
| 12 |
Hb_002025_190 |
0.1025645197 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 13 |
Hb_000999_060 |
0.1034020696 |
- |
- |
hypothetical protein POPTR_0018s11350g [Populus trichocarpa] |
| 14 |
Hb_003090_250 |
0.1038112733 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000429_080 |
0.1040367824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_009851_120 |
0.1045763819 |
- |
- |
PREDICTED: RING-H2 zinc finger protein RHA4a [Jatropha curcas] |
| 17 |
Hb_000754_010 |
0.108595514 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_024066_030 |
0.1090801052 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 19 |
Hb_002837_150 |
0.1101842891 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 20 |
Hb_001343_060 |
0.1105814463 |
- |
- |
Cytochrome P450 [Theobroma cacao] |