| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
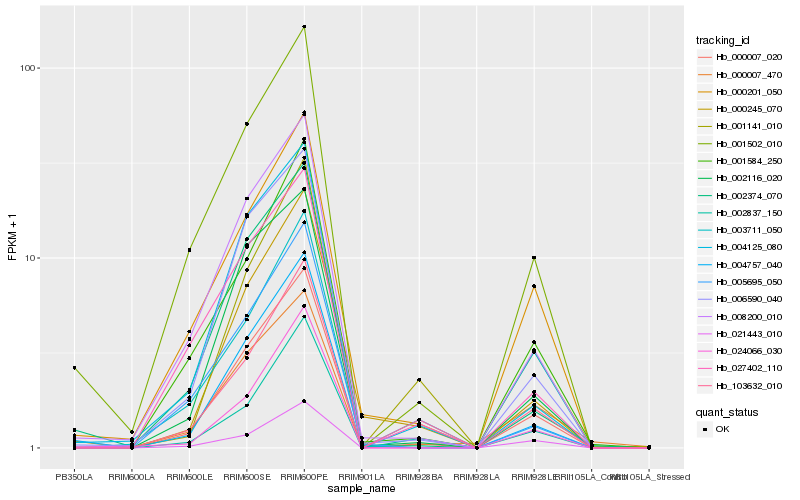
Hb_000007_020 |
0.0 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 2 |
Hb_008200_010 |
0.0817541603 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_006590_040 |
0.0826309661 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 4 |
Hb_001584_250 |
0.0862746374 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 5 |
Hb_001502_010 |
0.0911228495 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 6 |
Hb_002116_020 |
0.099770858 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 7 |
Hb_000201_050 |
0.1032188381 |
- |
- |
- |
| 8 |
Hb_021443_010 |
0.1040407751 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 9 |
Hb_004125_080 |
0.104981327 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Populus euphratica] |
| 10 |
Hb_027402_110 |
0.1103210579 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 11 |
Hb_103632_010 |
0.1107812399 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 12 |
Hb_024066_030 |
0.1128204472 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 13 |
Hb_005695_050 |
0.1134078379 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 14 |
Hb_000245_070 |
0.1149292909 |
- |
- |
PREDICTED: uncharacterized protein LOC105141276 [Populus euphratica] |
| 15 |
Hb_002374_070 |
0.115392568 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_001141_010 |
0.117903039 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 17 |
Hb_000007_470 |
0.1182118829 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 18 |
Hb_002837_150 |
0.1184642547 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 19 |
Hb_004757_040 |
0.1186471487 |
- |
- |
- |
| 20 |
Hb_003711_050 |
0.1220543348 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |