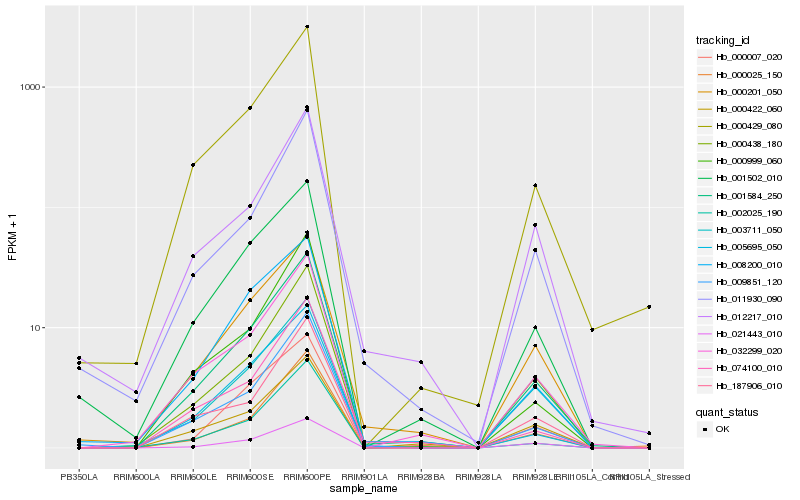
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001584_250 |
0.0 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 2 |
Hb_032299_020 |
0.069586983 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 3 |
Hb_000429_080 |
0.0762985106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000438_180 |
0.0782668044 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000201_050 |
0.0856022566 |
- |
- |
- |
| 6 |
Hb_009851_120 |
0.0857131207 |
- |
- |
PREDICTED: RING-H2 zinc finger protein RHA4a [Jatropha curcas] |
| 7 |
Hb_000007_020 |
0.0862746374 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 8 |
Hb_000025_150 |
0.0923774254 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 9 |
Hb_001502_010 |
0.0935769899 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 10 |
Hb_003711_050 |
0.0942055276 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 11 |
Hb_021443_010 |
0.0948911604 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 12 |
Hb_000422_060 |
0.0959332849 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 13 |
Hb_002025_190 |
0.0986822912 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 14 |
Hb_005695_050 |
0.098902272 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 15 |
Hb_074100_010 |
0.0995951846 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 16 |
Hb_011930_090 |
0.1000658382 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 17 |
Hb_187906_010 |
0.1013221689 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA2-like [Populus euphratica] |
| 18 |
Hb_008200_010 |
0.1020508631 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000999_060 |
0.1027346216 |
- |
- |
hypothetical protein POPTR_0018s11350g [Populus trichocarpa] |
| 20 |
Hb_012217_010 |
0.1062024806 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |